A CT-based deep learning radiomics nomogram for predicting the response to neoadjuvant chemotherapy in patients with locally advanced gastric cancer: A multicenter cohort study
- PMID: 35340629
- PMCID: PMC8943416
- DOI: 10.1016/j.eclinm.2022.101348
A CT-based deep learning radiomics nomogram for predicting the response to neoadjuvant chemotherapy in patients with locally advanced gastric cancer: A multicenter cohort study
Abstract
Background: Accurate prediction of treatment response to neoadjuvant chemotherapy (NACT) in individual patients with locally advanced gastric cancer (LAGC) is essential for personalized medicine. We aimed to develop and validate a deep learning radiomics nomogram (DLRN) based on pretreatment contrast-enhanced computed tomography (CT) images and clinical features to predict the response to NACT in patients with LAGC.
Methods: 719 patients with LAGC were retrospectively recruited from four Chinese hospitals between Dec 1st, 2014 and Nov 30th, 2020. The training cohort and internal validation cohort (IVC), comprising 243 and 103 patients, respectively, were randomly selected from center I; the external validation cohort1 (EVC1) comprised 207 patients from center II; and EVC2 comprised 166 patients from another two hospitals. Two imaging signatures, reflecting the phenotypes of the deep learning and handcrafted radiomics features, were constructed from the pretreatment portal venous-phase CT images. A four-step procedure, including reproducibility evaluation, the univariable analysis, the LASSO method, and the multivariable logistic regression analysis, was applied for feature selection and signature building. The integrated DLRN was then developed for the added value of the imaging signatures to independent clinicopathological factors for predicting the response to NACT. The prediction performance was assessed with respect to discrimination, calibration, and clinical usefulness. Kaplan-Meier survival curves based on the DLRN were used to estimate the disease-free survival (DFS) in the follow-up cohort (n = 300).
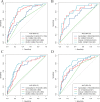
Findings: The DLRN showed satisfactory discrimination of good response to NACT and yielded the areas under the receiver operating curve (AUCs) of 0.829 (95% CI, 0.739-0.920), 0.804 (95% CI, 0.732-0.877), and 0.827 (95% CI, 0.755-0.900) in the internal and two external validation cohorts, respectively, with good calibration in all cohorts (p > 0.05). Furthermore, the DLRN performed significantly better than the clinical model (p < 0.001). Decision curve analysis confirmed that the DLRN was clinically useful. Besides, DLRN was significantly associated with the DFS of patients with LAGC (p < 0.05).
Interpretation: A deep learning-based radiomics nomogram exhibited a promising performance for predicting therapeutic response and clinical outcomes in patients with LAGC, which could provide valuable information for individualized treatment.
Keywords: AIC, Akaike information criterion; CT, computed tomography; DCA, decision curve analysis; DFS, disease free survival; DLRN, deep learning radiomics nomogram; Deep learning; GR, good response; ICC, interclass correlation coefficient; IDI, integrated discrimination improvement; LAGC, locally advanced gastric cancer; LASSO, least absolute shrinkage and selection operator; Locally advanced gastric cancer; NACT, neoadjuvant chemotherapy; NRI, Net reclassification index; Neoadjuvant chemotherapy; PR, poor response; ROC, Receiver operating characteristic; ROI, regions of interest; Radiomics nomogram; TRG, tumor regression grade.
© 2022 The Authors.
Conflict of interest statement
JR is an employee of GE Healthcare. YC received funding from National Natural Science Foundation of China (No. 82001789), China Postdoctoral Science Foundation (No. 2021M700897), and Project of Shanxi Provincial Health Commission (No. 2021XM51 and 2019058). ZL received funding from National Natural Science Foundation of China (No. 82001986), and Applied Basic Research Projects of Yunnan Province, China, Outstanding Youth Foundation (202101AW070001). YL received funding from National Natural Science Foundation of China (No. 82002702), and Youth Project of Natural Science Foundation of Hunan Science (No. 2020JJ5905). XY received funding from National Natural Science Foundation of China (No. 82171923), and Project of Shanxi Provincial Health Commission (No. 2020064). XG received funding from National Natural Science Foundation of China (No. 81871439). All other authors declare no competing interests.
Figures


Similar articles
-
A Deep Learning Radiomics Nomogram to Predict Response to Neoadjuvant Chemotherapy for Locally Advanced Cervical Cancer: A Two-Center Study.Diagnostics (Basel). 2023 Mar 11;13(6):1073. doi: 10.3390/diagnostics13061073. Diagnostics (Basel). 2023. PMID: 36980381 Free PMC article.
-
Ultrasound-based deep learning radiomics in the assessment of pathological complete response to neoadjuvant chemotherapy in locally advanced breast cancer.Eur J Cancer. 2021 Apr;147:95-105. doi: 10.1016/j.ejca.2021.01.028. Epub 2021 Feb 24. Eur J Cancer. 2021. PMID: 33639324
-
Deep learning nomogram for predicting neoadjuvant chemotherapy response in locally advanced gastric cancer patients.Abdom Radiol (NY). 2024 Nov;49(11):3780-3796. doi: 10.1007/s00261-024-04331-7. Epub 2024 May 26. Abdom Radiol (NY). 2024. PMID: 38796795 Free PMC article.
-
Deep learning or radiomics based on CT for predicting the response of gastric cancer to neoadjuvant chemotherapy: a meta-analysis and systematic review.Front Oncol. 2024 Mar 27;14:1363812. doi: 10.3389/fonc.2024.1363812. eCollection 2024. Front Oncol. 2024. PMID: 38601765 Free PMC article.
-
Diagnostic accuracy of radiomics-based machine learning for neoadjuvant chemotherapy response and survival prediction in gastric cancer patients: A systematic review and meta-analysis.Eur J Radiol. 2024 Apr;173:111249. doi: 10.1016/j.ejrad.2023.111249. Epub 2023 Dec 5. Eur J Radiol. 2024. PMID: 38382422
Cited by
-
A transformer-based deep learning model for early prediction of lymph node metastasis in locally advanced gastric cancer after neoadjuvant chemotherapy using pretreatment CT images.EClinicalMedicine. 2024 Aug 30;75:102805. doi: 10.1016/j.eclinm.2024.102805. eCollection 2024 Sep. EClinicalMedicine. 2024. PMID: 39281097 Free PMC article.
-
What benefit can be obtained from magnetic resonance imaging diagnosis with artificial intelligence in prostate cancer compared with clinical assessments?Mil Med Res. 2023 Jun 26;10(1):29. doi: 10.1186/s40779-023-00464-w. Mil Med Res. 2023. PMID: 37357263 Free PMC article. Review.
-
Computed tomography enterography-based deep learning radiomics to predict stratified healing in patients with Crohn's disease: a multicenter study.Insights Imaging. 2024 Nov 15;15(1):275. doi: 10.1186/s13244-024-01854-x. Insights Imaging. 2024. PMID: 39546153 Free PMC article.
-
The artificial intelligence revolution in gastric cancer management: clinical applications.Cancer Cell Int. 2025 Mar 21;25(1):111. doi: 10.1186/s12935-025-03756-4. Cancer Cell Int. 2025. PMID: 40119433 Free PMC article. Review.
-
Integrated radiomics and deep learning model for identifying medullary sponge kidney stones.Front Med (Lausanne). 2025 Jul 25;12:1623850. doi: 10.3389/fmed.2025.1623850. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40786086 Free PMC article.
References
-
- Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Al-Batran S.E., Homann N., Pauligk C., et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet. 2019;393:1948–1957. - PubMed
-
- Fazio N., Biffi R., Maibach R., et al. Preoperative versus postoperative docetaxel-cisplatin-fluorouracil (TCF) chemotherapy in locally advanced resectable gastric carcinoma: 10-year follow-up of the SAKK 43/99 phase III trial. Ann Oncol. 2016;27:668–673. - PubMed
-
- Lorenzen S., Blank S., Lordick F., Siewert J.R., Ott K. Prediction of response and prognosis by a score including only pretherapeutic parameters in 410 neoadjuvant treated gastric cancer patients. Ann Surg Oncol. 2012;19:2119–2127. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

