The dynamics of circulating SARS-CoV-2 lineages in Bogor and surrounding areas reflect variant shifting during the first and second waves of COVID-19 in Indonesia
- PMID: 35341058
- PMCID: PMC8953504
- DOI: 10.7717/peerj.13132
The dynamics of circulating SARS-CoV-2 lineages in Bogor and surrounding areas reflect variant shifting during the first and second waves of COVID-19 in Indonesia
Abstract
Background: Indonesia is one of the Southeast Asian countries with high case numbers of COVID-19 with up to 4.2 million confirmed cases by 29 October 2021. Understanding the genome of SARS-CoV-2 is crucial for delivering public health intervention as certain variants may have different attributes that can potentially affect their transmissibility, as well as the performance of diagnostics, vaccines, and therapeutics.
Objectives: We aimed to investigate the dynamics of circulating SARS-CoV-2 variants over a 15-month period in Bogor and its surrounding areas in correlation with the first and second wave of COVID-19 in Indonesia.
Methods: Nasopharyngeal and oropharyngeal swab samples collected from suspected patients from Bogor, Jakarta and Tangerang were confirmed for SARS-CoV-2 infection with RT-PCR. RNA samples of those confirmed patients were subjected to whole genome sequencing using the ARTIC Network protocol and sequencer platform from Oxford Nanopore Technologies (ONT).
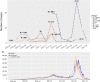
Results: We successfully identified 16 lineages and six clades out of 202 samples (male n = 116, female n = 86). Genome analysis revealed that Indonesian lineage B.1.466.2 dominated during the first wave (n = 48, 23.8%) while Delta variants (AY.23, AY.24, AY.39, AY.42, AY.43 dan AY.79) were dominant during the second wave (n = 53, 26.2%) following the highest number of confirmed cases in Indonesia. In the spike protein gene, S_D614G and S_P681R changes were dominant in both B.1.466.2 and Delta variants, while N439K was only observed in B.1.466.2 (n = 44) and B.1.470 (n = 1). Additionally, the S_T19R, S_E156G, S_F157del, S_R158del, S_L452R, S_T478K, S_D950N and S_V1264L changes were only detected in Delta variants, consistent with those changes being characteristic of Delta variants in general.
Conclusions: We demonstrated a shift in SARS-CoV-2 variants from the first wave of COVID-19 to Delta variants in the second wave, during which the number of confirmed cases surpassed those in the first wave of COVID-19 pandemic. Higher proportion of unique mutations detected in Delta variants compared to the first wave variants indicated potential mutational effects on viral transmissibility that correlated with a higher incidence of confirmed cases. Genomic surveillance of circulating variants, especially those with higher transmissibility, should be continuously conducted to rapidly inform decision making and support outbreak preparedness, prevention, and public health response.
Keywords: Bogor; COVID-19; Delta variants; Genomic surveillance; Indonesian lineages; Nanopore; SARS-CoV-2; Variant shifting.
©2022 Prasetyoputri et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures




References
-
- Allen H, Vusirikala A, Flannagan J, Twohig KA, Zaidi A, Chudasama D, Lamagni T, Groves N, Turner C, Rawlinson C, Lopez-Bernal J, Harris R, Charlett A, Dabrera G, Kall M, the COVID-19 Genomics UK (COG-UK Consortium) Household transmission of COVID-19 cases associated with SARS-CoV-2 delta variant (B.1.617.2): national case-control study. The Lancet Regional Health—Europe. 2021;12:100252. - PMC - PubMed
-
- Baker DJ, Aydin A, Le-Viet T, Kay GL, Rudder S, Martins LDO, Tedim AP, Kolyva A, Diaz M, Alikhan N-F, Meadows L, Bell A, Gutierrez AV, Trotter AJ, Thomson NM, Gilroy R, Griffith L, Adriaenssens EM, Stanley R, Charles IG, Elumogo N, Wain J, Prakash R, Meader E, Mather AE, Webber MA, Dervisevic S, Page AJ, OGrady J. CoronaHiT: High throughput sequencing of SARS-CoV-2 genomes. bioRxiv. 2021 2020.06.24.162156. - PMC - PubMed
-
- BNPB Situasi COVID-19 di Indonesia. 2021. https://experience.arcgis.com/experience/57237ebe9c5b4b1caa1b93e79c920338. [13 May 2021]. https://experience.arcgis.com/experience/57237ebe9c5b4b1caa1b93e79c920338
-
- Bull RA, Adikari TN, Ferguson JM, Hammond JM, Stevanovski I, Beukers AG, Naing Z, Yeang M, Verich A, Gamaarachchi H, Kim KW, Luciani F, Stelzer-Braid S, Eden J-S, Rawlinson WD, Van Hal SJ, Deveson IW. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nature Communications. 2020;11(1):6272. doi: 10.1038/s41467-020-20075-6. - DOI - PMC - PubMed
-
- CDC SARS-CoV-2 variant classifications and definitions. 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html. [October 2021]. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

