Identifying and predicting amyotrophic lateral sclerosis clinical subgroups: a population-based machine-learning study
- PMID: 35341712
- PMCID: PMC9038712
- DOI: 10.1016/S2589-7500(21)00274-0
Identifying and predicting amyotrophic lateral sclerosis clinical subgroups: a population-based machine-learning study
Abstract
Background: Amyotrophic lateral sclerosis (ALS) is known to represent a collection of overlapping syndromes. Various classification systems based on empirical observations have been proposed, but it is unclear to what extent they reflect ALS population substructures. We aimed to use machine-learning techniques to identify the number and nature of ALS subtypes to obtain a better understanding of this heterogeneity, enhance our understanding of the disease, and improve clinical care.
Methods: In this retrospective study, we applied unsupervised Uniform Manifold Approximation and Projection [UMAP]) modelling, semi-supervised (neural network UMAP) modelling, and supervised (ensemble learning based on LightGBM) modelling to a population-based discovery cohort of patients who were diagnosed with ALS while living in the Piedmont and Valle d'Aosta regions of Italy, for whom detailed clinical data, such as age at symptom onset, were available. We excluded patients with missing Revised ALS Functional Rating Scale (ALSFRS-R) feature values from the unsupervised and semi-supervised steps. We replicated our findings in an independent population-based cohort of patients who were diagnosed with ALS while living in the Emilia Romagna region of Italy.
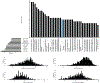
Findings: Between Jan 1, 1995, and Dec 31, 2015, 2858 patients were entered in the discovery cohort. After excluding 497 (17%) patients with missing ALSFRS-R feature values, data for 42 clinical features across 2361 (83%) patients were available for the unsupervised and semi-supervised analysis. We found that semi-supervised machine learning produced the optimum clustering of the patients with ALS. These clusters roughly corresponded to the six clinical subtypes defined by the Chiò classification system (ie, bulbar, respiratory, flail arm, classical, pyramidal, and flail leg ALS). Between Jan 1, 2009, and March 1, 2018, 1097 patients were entered in the replication cohort. After excluding 108 (10%) patients with missing ALSFRS-R feature values, data for 42 clinical features across 989 patients were available for the unsupervised and semi-supervised analysis. All 1097 patients were included in the supervised analysis. The same clusters were identified in the replication cohort. By contrast, other ALS classification schemes, such as the El Escorial categories, Milano-Torino clinical staging, and King's clinical stages, did not adequately label the clusters. Supervised learning identified 11 clinical parameters that predicted ALS clinical subtypes with high accuracy (area under the curve 0·982 [95% CI 0·980-0·983]).
Interpretation: Our data-driven study provides insight into the ALS population substructure and confirms that the Chiò classification system successfully identifies ALS subtypes. Additional validation is required to determine the accuracy and clinical use of these algorithms in assigning clinical subtypes. Nevertheless, our algorithms offer a broad insight into the clinical heterogeneity of ALS and help to determine the actual subtypes of disease that exist within this fatal neurodegenerative syndrome. The systematic identification of ALS subtypes will improve clinical care and clinical trial design.
Funding: US National Institute on Aging, US National Institutes of Health, Italian Ministry of Health, European Commission, University of Torino Rita Levi Montalcini Department of Neurosciences, Emilia Romagna Regional Health Authority, and Italian Ministry of Education, University, and Research.
Translations: For the Italian and German translations of the abstract see Supplementary Materials section.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests BJT holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 (patent numbers EP2751284A1, CA2846307A, and 20180187262); received research grants from the Myasthenia Gravis Foundation, ALS Association, US Center for Disease Control and Prevention, US Department of Veterans Affairs, MSD, and Cerevel Therapeutics; receives funding through the Intramural Research Program at the US National Institutes of Health (NIH), is on the scientific advisory committee of the American Neurological Association, is an associate editor of Brain, and is on the editorial boards of Journal of Neurology, Neurosurgery, and Psychiatry, Neurobiology of Aging, and eClinicalMedicine. JM received research grants from the Fondazione Italiana di Ricerca per la Sclerosi Laterale Amiotrofica, Agenzia Italiana del Farmaco, Italian Ministry of Health, Emilia Romagna Regional Health Authority, and Pfizer. ACh received research funding and honoraria for lectures from Biogen; sits on advisory boards for Mitsubishi Tanabe Pharma, Roche, Denali Therapeutics, Cytokinetics, Biogen, Amylyx Pharmaceuticals, and Sanofi; and participates in data safety monitoring boards for Lilly and AB Science. RV received research scholarship funding from the Rotary Club (global grant GG2094854). FF is employed by Data Tecnica International. MAN is employed by Data Tecnica International and is an adviser for Clover Therapeutics and Neuron23. AD is employed by Data Tecnica International. All other authors declare no competing interests.
Figures
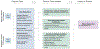
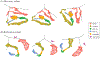
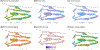

References
-
- Hirtz D, Thurman DJ, Gwinn-Hardy K, Mohamed M, Chaudhuri AR, Zalutsky R. How common are the “common” neurologic disorders? Neurology 2007; 68: 326–37 - PubMed
-
- Byrne S, Bede P, Elamin M, et al. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler 2011; 12: 157–9 - PubMed
-
- de Carvalho M, Dengler R, Eisen A, et al. Electrodiagnostic criteria for diagnosis of ALS. Clin Neurophysiol 2008; 119: 497–503 - PubMed
-
- Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci 1994; 124 Suppl: 96–107 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

