Molecular Virology of SARS-CoV-2 and Related Coronaviruses
- PMID: 35343760
- PMCID: PMC9199417
- DOI: 10.1128/mmbr.00026-21
Molecular Virology of SARS-CoV-2 and Related Coronaviruses
Abstract
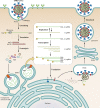
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The global COVID-19 pandemic continues to threaten the lives of hundreds of millions of people, with a severe negative impact on the global economy. Although several COVID-19 vaccines are currently being administered, none of them is 100% effective. Moreover, SARS-CoV-2 variants remain an important worldwide public health issue. Hence, the accelerated development of efficacious antiviral agents is urgently needed. Coronavirus depends on various host cell factors for replication. An ongoing research objective is the identification of host factors that could be exploited as targets for drugs and compounds effective against SARS-CoV-2. In the present review, we discuss the molecular mechanisms of SARS-CoV-2 and related coronaviruses, focusing on the host factors or pathways involved in SARS-CoV-2 replication that have been identified by genome-wide CRISPR screening.
Keywords: SARS-CoV-2; coronavirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Gao YD, Ding M, Dong X, Zhang JJ, Kursat Azkur A, Azkur D, Gan H, Sun YL, Fu W, Li W, Liang HL, Cao YY, Yan Q, Cao C, Gao HY, Bruggen MC, van de Veen W, Sokolowska M, Akdis M, Akdis CA. 2021. Risk factors for severe and critically ill COVID-19 patients: a review. Allergy 76:428–455. 10.1111/all.14657. - DOI - PubMed
-
- Gold JAW, Rossen LM, Ahmad FB, Sutton P, Li Z, Salvatore PP, Coyle JP, DeCuir J, Baack BN, Durant TM, Dominguez KL, Henley SJ, Annor FB, Fuld J, Dee DL, Bhattarai A, Jackson BR. 2020. Race, ethnicity, and age trends in persons who died from COVID-19—United States, May-August 2020. MMWR Morb Mortal Wkly Rep 69:1517–1521. 10.15585/mmwr.mm6942e1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

