Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity
- PMID: 35343766
- PMCID: PMC9044946
- DOI: 10.1128/jvi.00128-22
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity
Erratum in
-
Erratum for Essalmani et al., "Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity".J Virol. 2022 Jul 13;96(13):e0074522. doi: 10.1128/jvi.00745-22. Epub 2022 Jun 6. J Virol. 2022. PMID: 35658532 Free PMC article. No abstract available.
Abstract
The spike protein (S) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directs infection of the lungs and other tissues following its binding to the angiotensin-converting enzyme 2 (ACE2) receptor. For effective infection, the S protein is cleaved at two sites: S1/S2 and S2'. The "priming" of the surface S protein at S1/S2 (P
Keywords: Calu-3 cells; HeLa cells; SARS-CoV-2; TMPRSS2; cell-to-cell fusion; furin; spike; viral entry; viral infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


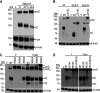


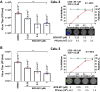
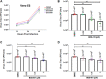
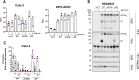




References
MeSH terms
Substances
Grants and funding
- 148363/Gouvernement du Canada | Canadian Institutes of Health Research (IRSC)
- 154324/Gouvernement du Canada | Canadian Institutes of Health Research (IRSC)
- 950-231335/Gouvernement du Canada | Canadian Institutes of Health Research (IRSC)
- HAL 157986/Gouvernement du Canada | Canadian Institutes of Health Research (IRSC)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

