Indirubin-3'-monoxime acts as proteasome inhibitor: Therapeutic application in multiple myeloma
- PMID: 35344764
- PMCID: PMC8958548
- DOI: 10.1016/j.ebiom.2022.103950
Indirubin-3'-monoxime acts as proteasome inhibitor: Therapeutic application in multiple myeloma
Abstract
Background: Multiple myeloma (MM) is still an incurable malignancy of plasma cells. Proteasome inhibitors (PIs) work as the backbone agent and have greatly improved the outcome in majority of newly diagnosed patients with myeloma. However, drug resistance remains the major obstacle causing treatment failure in clinical practice. Here, we investigated the effects of Indirubin-3'-monoxime (I3MO), one of the derivatives of Indirubin, in the treatment of MM.
Methods: MM patient primary samples and human cell lines were examined. I3MO effects on myeloma treatment and the underling molecular mechanisms were investigated via in vivo and in vitro study.
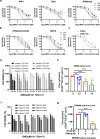
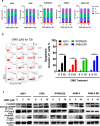
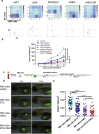
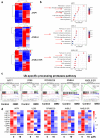
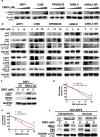
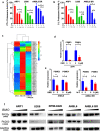
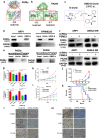
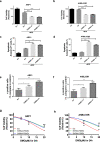
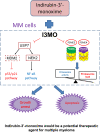
Findings: Our results demonstrated the anti-MM activity of I3MO in both drug- sensitive and -resistance MM cells. I3MO sensitizes MM cells to bortezomib-induced apoptosis. Mechanistically, I3MO acts as a multifaceted regulator of cell death, which induced DNA damage, cell cycle arrest, and abrogates NF-κB activation. I3MO efficiently down-regulated USP7 expression, promoted NEK2 degradation, and suppressed NF-κB signaling in MM. Our study reported that I3MO directly bound with and caused the down-regulation of PA28γ (PSME3), and PA200 (PSME4), the proteasome activators. Knockdown of PSME3 or PSME4 caused the inhibition of proteasome capacity and the overload of paraprotein, which sensitizes MM cells to bortezomib-mediated growth arrest. Clinical data demonstrated that PSME3 and PSME4 are over-expressed in relapsed/refractory MM (RRMM) and associated with inferior outcome.
Interpretation: Altogether, our study indicates that I3MO is agent triggering proteasome inhibition and represents a promising therapeutic strategy to improve patient outcome in MM.
Fundings: A full list of funding can be found in the acknowledgements.
Keywords: Indirubin-3’-monoxime (I3MO); Multiple myeloma; PSME3 (PA28γ); PSME4 (PA200); Proteasome inhibition.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Dr. Kenneth. C. Anderson is the consultant: Pfizer, Amgen, Astrazeneca, Janssen, Precision Biosciences, Mana and Window. Founder/Stock Shareholder: C4 Therapeutics, Oncopep, Raqia and NextRNA. Other authors declared no competing financial interests in relation to the work described.
Figures

References
-
- Kumar S.K., Anderson K.C. Immune Therapies in Multiple Myeloma. Clin Cancer Res. 2016;22(22):5453–5460. - PubMed
-
- An G., Yan Y., Xu Y., et al. Monitoring the cytogenetic architecture of minimal residual plasma cells indicates therapy-induced clonal selection in multiple myeloma. Leukemia. 2020;34(2):578–588. - PubMed
-
- Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12(9):1191–1197. - PubMed
-
- Adams J. The proteasome: structure, function, and role in the cell. Cancer Treat Rev. 2003;29:3–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

