Statistical inference for a quasi birth-death model of RNA transcription
- PMID: 35346020
- PMCID: PMC8961911
- DOI: 10.1186/s12859-022-04638-6
Statistical inference for a quasi birth-death model of RNA transcription
Abstract
Background: A birth-death process of which the births follow a hypoexponential distribution with L phases and are controlled by an on/off mechanism, is a population process which we call the on/off-seq-L process. It is a suitable model for the dynamics of a population of RNA molecules in a single living cell. Motivated by this biological application, our aim is to develop a statistical method to estimate the model parameters of the on/off-seq-L process, based on observations of the population size at discrete time points, and to apply this method to real RNA data.
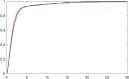
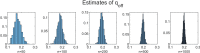
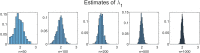
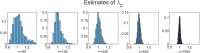
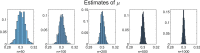
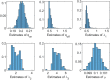
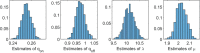
Methods: It is shown that the on/off-seq-L process can be seen as a quasi birth-death process, and an Erlangization technique can be used to approximate the corresponding likelihood function. An extensive simulation-based numerical study is carried out to investigate the performance of the resulting estimation method.
Results and conclusion: A statistical method is presented to find maximum likelihood estimates of the model parameters for the on/off-seq-L process. Numerical complications related to the likelihood maximization are identified and analyzed, and solutions are presented. The proposed estimation method is a highly accurate method to find the parameter estimates. Based on real RNA data, the on/off-seq-3 process emerges as the best model to describe RNA transcription.
Keywords: Erlangization technique; Maximum likelihood estimation; Quasi birth–death process; RNA transcription.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

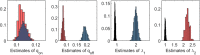



References
-
- Asmussen S, Avram F, Usabel M. Erlangian Approximations for Finite-Horizon Ruin Probabilities. ASTIN Bull. 2002;32:267–281. doi: 10.2143/AST.32.2.1029. - DOI
-
- Bright L, Taylor P. Calculating the equilibrium distribution in level dependent Quasi-Birth-and-Death processes. Stoch Model. 1995;11:497–526. doi: 10.1080/15326349508807357. - DOI
-
- Kandhavelu M, Mannerström H, Gupta A, Häkkinen A, Lloyd-Price J, Yli-Harja O, Ribeiro AS. In vivo kinetics of transcription initiation of the lar promoter in Escherichia coli. Evidence for a sequential mechanism with two rate-limiting steps. BMC Syst Biol. 2011;5:1–9. doi: 10.1186/1752-0509-5-149. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

