SHOOT: phylogenetic gene search and ortholog inference
- PMID: 35346327
- PMCID: PMC8962542
- DOI: 10.1186/s13059-022-02652-8
SHOOT: phylogenetic gene search and ortholog inference
Abstract
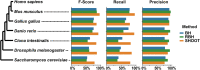
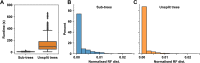
Determining the evolutionary relationships between genes is fundamental to comparative biological research. Here, we present SHOOT. SHOOT searches a user query sequence against a database of phylogenetic trees and returns a tree with the query sequence correctly placed within it. We show that SHOOT performs this analysis with comparable speed to a BLAST search. We demonstrate that SHOOT phylogenetic placements are as accurate as conventional tree inference, and it can identify orthologs with high accuracy. In summary, SHOOT is a fast and accurate tool for phylogenetic analyses of novel query sequences. It is available online at www.shoot.bio .
Keywords: Orthology inference; Phylogenetic tree inference; Sequence similarity search.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

