Molecular rhythm alterations in prefrontal cortex and nucleus accumbens associated with opioid use disorder
- PMID: 35347109
- PMCID: PMC8960783
- DOI: 10.1038/s41398-022-01894-1
Molecular rhythm alterations in prefrontal cortex and nucleus accumbens associated with opioid use disorder
Abstract
Severe and persistent disruptions to sleep and circadian rhythms are common in people with opioid use disorder (OUD). Preclinical evidence suggests altered molecular rhythms in the brain modulate opioid reward and relapse. However, whether molecular rhythms are disrupted in the brains of people with OUD remained an open question, critical to understanding the role of circadian rhythms in opioid addiction. Using subjects' times of death as a marker of time of day, we investigated transcriptional rhythms in the brains of subjects with OUD compared to unaffected comparison subjects. We discovered rhythmic transcripts in both the dorsolateral prefrontal cortex (DLPFC) and nucleus accumbens (NAc), key brain areas involved in OUD, that were largely distinct between OUD and unaffected subjects. Fewer rhythmic transcripts were identified in DLPFC of subjects with OUD compared to unaffected subjects, whereas in the NAc, nearly double the number of rhythmic transcripts was identified in subjects with OUD. In NAc of subjects with OUD, rhythmic transcripts peaked either in the evening or near sunrise, and were associated with an opioid, dopamine, and GABAergic neurotransmission. Associations with altered neurotransmission in NAc were further supported by co-expression network analysis which identified OUD-specific modules enriched for transcripts involved in dopamine, GABA, and glutamatergic synaptic functions. Additionally, rhythmic transcripts in DLPFC and NAc of subjects with OUD were enriched for genomic loci associated with sleep-related GWAS traits, including sleep duration and insomnia. Collectively, our findings connect transcriptional rhythm changes in opioidergic, dopaminergic, GABAergic signaling in the human brain to sleep-related traits in opioid addiction.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
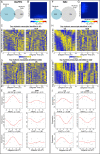




References
-
- Gossop M, Green L, Phillips G, Bradley B. Lapse, relapse and survival among opiate addicts after treatment. A prospective follow-up study. Br J Psychiatry. 1989;154:348–53. - PubMed
-
- Garcia AN, Salloum IM. Polysomnographic sleep disturbances in nicotine, caffeine, alcohol, cocaine, opioid, and cannabis use: a focused review. Am J Addict. 2015;24:590–8. - PubMed
-
- Cao M, Javaheri S. Effects of chronic opioid use on sleep and wake. Sleep Med Clin. 2018;13:271–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01HL150432/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- F30DA053020/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- R01 DA051390/DA/NIDA NIH HHS/United States
- DP1 DA046585/DA/NIDA NIH HHS/United States
- F30 DA053020/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

