Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma
- PMID: 35347134
- PMCID: PMC8960779
- DOI: 10.1038/s41467-022-29164-0
Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma
Erratum in
-
Publisher Correction: Single-cell transcriptomic analysis suggests two molecularly distinct subtypes of intrahepatic cholangiocarcinoma.Nat Commun. 2022 May 17;13(1):2848. doi: 10.1038/s41467-022-30599-8. Nat Commun. 2022. PMID: 35581192 Free PMC article. No abstract available.
Abstract
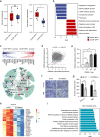
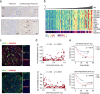
Intrahepatic cholangiocarcinoma (iCCA) is a highly heterogeneous cancer with limited understanding of its classification and tumor microenvironment. Here, by performing single-cell RNA sequencing on 144,878 cells from 14 pairs of iCCA tumors and non-tumor liver tissues, we find that S100P and SPP1 are two markers for iCCA perihilar large duct type (iCCAphl) and peripheral small duct type (iCCApps). S100P + SPP1- iCCAphl has significantly reduced levels of infiltrating CD4+ T cells, CD56+ NK cells, and increased CCL18+ macrophages and PD1+CD8+ T cells compared to S100P-SPP1 + iCCApps. The transcription factor CREB3L1 is identified to regulate the S100P expression and promote tumor cell invasion. S100P-SPP1 + iCCApps has significantly more SPP1+ macrophage infiltration, less aggressiveness and better survival than S100P + SPP1- iCCAphl. Moreover, S100P-SPP1 + iCCApps harbors tumor cells at different status of differentiation, such as ALB + hepatocyte differentiation and ID3+ stemness. Our study extends the understanding of the diversity of tumor cells in iCCA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Nakanuma, Y., Klimstra, D., Komuta, M. & Zen, Y. in WHO Classification of Tumours: Digestive System Tumours 5th edn (ed. WHO Classification of Tumors Editorial Board.) 8, 254–259 (World Health Organization, 2019).
-
- Kendall T, et al. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019;39:7–18. - PubMed
-
- Aishima S, Oda Y. Pathogenesis and classification of intrahepatic cholangiocarcinoma: different characters of perihilar large duct type versus peripheral small duct type. J. Hepatobiliary Pancreat. Sci. 2015;22:94–100. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

