Antimicrobial resistance determinants in silage
- PMID: 35347213
- PMCID: PMC8960843
- DOI: 10.1038/s41598-022-09296-5
Antimicrobial resistance determinants in silage
Abstract
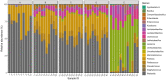
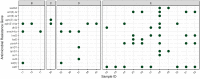
Animal products may play a role in developing and spreading antimicrobial resistance in several ways. On the one hand, residues of antibiotics not adequately used in animal farming can enter the human body via food. However, resistant bacteria may also be present in animal products, which can transfer the antimicrobial resistance genes (ARG) to the bacteria in the consumer's body by horizontal gene transfer. As previous studies have shown that fermented foods have a meaningful ARG content, it is indicated that such genes may also be present in silage used as mass feed in the cattle sector. In our study, we aspired to answer what ARGs occur in silage and what mobility characteristics they have? For this purpose, we have analyzed bioinformatically 52 freely available deep sequenced silage samples from shotgun metagenome next-generation sequencing. A total of 16 perfect matched ARGs occurred 54 times in the samples. More than half of these ARGs are mobile because they can be linked to integrative mobile genetic elements, prophages or plasmids. Our results point to a neglected but substantial ARG source in the food chain.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Eastridge M. Major advances in applied dairy cattle nutrition. J. Dairy Sci. 2006;89:1311–1323. - PubMed
-
- Driehuis F, Spanjer M, Scholten J, Te Giffel M. Occurrence of mycotoxins in maize, grass and wheat silage for dairy cattle in the Netherlands. Food Addit. Contam. 2008;1:41–50. - PubMed
-
- Cabezón E, Ripoll-Rozada J, Peña A, De La Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015;39:81–95. - PubMed
-
- Goh, S. Clostridium difficile: Methods and Protocols, chap. Phage Transduction, 177–185 (Springer, 2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

