HYBRiD: hydrogel-reinforced DISCO for clearing mammalian bodies
- PMID: 35347322
- PMCID: PMC9337799
- DOI: 10.1038/s41592-022-01427-0
HYBRiD: hydrogel-reinforced DISCO for clearing mammalian bodies
Abstract
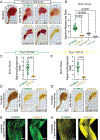
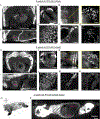
The recent development of solvent- and polymer-based brain-clearing techniques has advanced our ability to visualize the mammalian nervous system in three dimensions. However, it remains challenging to image the mammalian body en bloc. Here we developed HYBRiD (hydrogel-based reinforcement of three-dimensional imaging solvent-cleared organs (DISCO)), by recombining components of organic- and polymer-based clearing pipelines. We achieved high transparency and protein retention, as well as compatibility with direct fluorescent imaging and immunostaining in cleared mammalian bodies. Using parvalbumin- and somatostatin-Cre models, we demonstrated the utility of HYBRiD for whole-body imaging of genetically encoded fluorescent reporters without antibody enhancement of signals in newborn and juvenile mice. Using K18-hACE2 transgenic mice, HYBRiD enabled perfusion-free clearing and visualization of SARS-CoV-2 infection in a whole mouse chest, revealing macroscopic and microscopic features of viral pathology in the same sample. HYBRiD offers a simple and universal solution to visualize large heterogeneous body parts or entire animals for basic and translational research.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures









References
METHOD REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

