Application of nanopore sequencing in diagnosis of secondary infections in patients with severe COVID-19
- PMID: 35347908
- PMCID: PMC8931600
- DOI: 10.3724/zdxbyxb-2021-0158
Application of nanopore sequencing in diagnosis of secondary infections in patients with severe COVID-19
Abstract
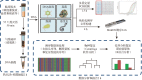
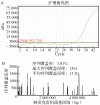
To explore the application value of nanopore sequencing technique in the diagnosis and treatment of secondary infections in patients with severe coronavirus disease 2019 (COVID-19). A total of 77 clinical specimens from 3 patients with severe COVID-19 were collected. After heat inactivation, all samples were subjected to total nucleic acid extraction based on magnetic bead enrichment. The extracted DNA was used for DNA library construction, then nanopore real-time sequencing detection was performed. The sequencing data were subjected to Centrifuge software database species matching and R program differential analysis to obtain potential pathogen identification. Nanopore sequencing results were compared with respiratory pathogen qPCR panel screening and conventional microbiological testing results to verify the effectiveness of nanopore sequencing detection. Nanopore sequencing results showed that positive pathogen were obtained in 44 specimens (57.1%). The potential pathogens identified by nanopore sequencing included , , and , et al. , , were also detected in clinical microbiological culture-based detection; was detected in respiratory pathogen screening qPCR panel; was only detected by the nanopore sequencing technique. Comprehensive considerations with the clinical symptoms, the patient was treated with antibiotics against , and the infection was controlled. Nanopore sequencing may assist the diagnosis and treatment of severe COVID-19 patients through rapid identification of potential pathogens.
Keywords: Coronavirus disease 2019; Metagenomics next-generation sequencing; Nanopore sequencing; Pathogen detection; Severe acute respiratory syndrome coronavirus 2.
Conflict of interest statement
所有作者均声明不存在利益冲突
Figures
References
-
- BERLIN D A, GULICK R M, MARTINEZ F J. Severe COVID-19[J] N Engl J Med. . 2020;383(25):2451–2460. doi: 10.1056/NEJMcp2009575. - DOI - PubMed
-
- YANG S, ROTHMAN R E. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings[J] Lancet Infect Dis. . 2004;4(6):337–348. doi: 10.1016/S1473-3099(04)01044-8. - DOI - PMC - PubMed
-
- 中华医学会检验医学分会. 高通量宏基因组测序技术检测病原微生物的临床应用规范化专家共识[J]. 中华检验医学杂志, 2021, 43(12): 1181-1195
- Chinese Society of Laboratory Medicine. Expert consensus on clinical standardized application of metagenomics next‐generation sequencing for detection of pathogenic microorganisms[J]. Chinese Journal of Laboratory Medicine, 2021, 43(12): 1181-1195. (in Chinese)
-
- 孙波, 郑光辉, 高阳, 等. 宏基因组技术在感染性疾病中的应用[J]. 中国临床新医学, 2021, 14(1): 19-22
- SUN Bo, ZHENG Guanghui, GAO Yang, et al. Application of metagenome technology in infectious diseases[J]. Chinese Journal of New Clinical Medicine, 2021, 14(1): 19-22. (in Chinese)
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

