Capturing an Early Gene Induction Event during Wood Decay by the Brown Rot Fungus Rhodonia placenta
- PMID: 35348388
- PMCID: PMC9040566
- DOI: 10.1128/aem.00188-22
Capturing an Early Gene Induction Event during Wood Decay by the Brown Rot Fungus Rhodonia placenta
Abstract
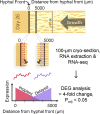
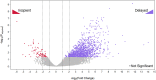
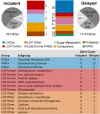
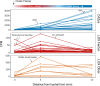
Brown rot fungi dominate wood decomposition in coniferous forests, and their carbohydrate-selective mechanisms are of commercial interest. Brown rot was recently described as a two-step, sequential mechanism orchestrated by fungi using differentially expressed genes (DEGs) and consisting of oxidation via reactive oxygen species (ROS) followed by enzymatic saccharification. There have been indications, however, that the initial oxidation step itself might require induction. To capture this early gene regulation event, here, we integrated fine-scale cryosectioning with whole-transcriptome sequencing to dissect gene expression at the single-hyphal-cell scale (tens of micrometers). This improved the spatial resolution 50-fold, relative to previous work, and we were able to capture the activity of the first 100 μm of hyphal front growth by Rhodonia placenta in aspen wood. This early decay period was dominated by delayed gene expression patterns as the fungus ramped up its mechanism. These delayed DEGs included many genes implicated in ROS pathways (lignocellulose oxidation [LOX]) that were previously and incorrectly assumed to be constitutively expressed. These delayed DEGs, which include those with and without predicted functions, also create a focused subset of target genes for functional genomics. However, this delayed pattern was not universal, with a few genes being upregulated immediately at the hyphal front. Most notably, this included a gene commonly implicated in hydroquinone and iron redox cycling: benzoquinone reductase. IMPORTANCE Earth's aboveground terrestrial biomass is primarily wood, and fungi dominate wood decomposition. Here, we studied these fungal pathways in a common "brown rot"-type fungus, Rhodonia placenta, that selectively extracts sugars from carbohydrates embedded within wood lignin. Using a space-for-time design to map fungal gene expression at the extreme hyphal front in wood, we made two discoveries. First, we found that many genes long assumed to be "on" (constitutively expressed) from the very beginning of decay were instead "off" before being upregulated, when mapped (via transcriptome sequencing [RNA-seq]) at a high resolution. Second, we found that the gene encoding benzoquinone reductase was "on" in incipient decay and quickly downregulated, implying a key role in "kick-starting" brown rot.
Keywords: RNA; biodegradation; decomposition; lignocellulose; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A Fungal Secretome Adapted for Stress Enabled a Radical Wood Decay Mechanism.mBio. 2021 Aug 31;12(4):e0204021. doi: 10.1128/mBio.02040-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399614 Free PMC article.
-
Gene Regulation Shifts Shed Light on Fungal Adaption in Plant Biomass Decomposers.mBio. 2019 Nov 19;10(6):e02176-19. doi: 10.1128/mBio.02176-19. mBio. 2019. PMID: 31744914 Free PMC article.
-
Oxidative Damage Control during Decay of Wood by Brown Rot Fungus Using Oxygen Radicals.Appl Environ Microbiol. 2018 Oct 30;84(22):e01937-18. doi: 10.1128/AEM.01937-18. Print 2018 Nov 15. Appl Environ Microbiol. 2018. PMID: 30194102 Free PMC article.
-
Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin.Int Microbiol. 2005 Sep;8(3):195-204. Int Microbiol. 2005. PMID: 16200498 Review.
-
Lignin-modifying enzymes in filamentous basidiomycetes--ecological, functional and phylogenetic review.J Basic Microbiol. 2010 Feb;50(1):5-20. doi: 10.1002/jobm.200900338. J Basic Microbiol. 2010. PMID: 20175122 Review.
Cited by
-
Gene Expression by a Model Fungus in the Ascomycota Provides Insight Into the Decay of Fungal Necromass.Environ Microbiol. 2024 Dec;26(12):e70006. doi: 10.1111/1462-2920.70006. Environ Microbiol. 2024. PMID: 39647917 Free PMC article.
-
A Laccase Gene Reporting System That Enables Genetic Manipulations in a Brown Rot Wood Decomposer Fungus Gloeophyllum trabeum.Microbiol Spectr. 2023 Feb 14;11(1):e0424622. doi: 10.1128/spectrum.04246-22. Epub 2023 Jan 18. Microbiol Spectr. 2023. PMID: 36651769 Free PMC article.
-
Micromorphological features of brown rotted wood revealed by broad argon ion beam milling.Sci Rep. 2024 Dec 30;14(1):32003. doi: 10.1038/s41598-024-83578-y. Sci Rep. 2024. PMID: 39738786 Free PMC article.
-
Wood decay under anoxia by the brown-rot fungus Fomitopsis pinicola.Nat Commun. 2025 Aug 9;16(1):7352. doi: 10.1038/s41467-025-62567-3. Nat Commun. 2025. PMID: 40783394 Free PMC article.
-
Unlocking the distinctive enzymatic functions of the early plant biomass deconstructive genes in a brown rot fungus by cell-free protein expression.Appl Environ Microbiol. 2024 May 21;90(5):e0012224. doi: 10.1128/aem.00122-24. Epub 2024 Apr 3. Appl Environ Microbiol. 2024. PMID: 38567954 Free PMC article.
References
-
- Houghton RA, Hall F, Goetz SJ. 2009. Importance of biomass in the global carbon cycle. J Geophys Res 114:G00E03.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

