Identification of Functional Heterogeneity of Carcinoma-Associated Fibroblasts with Distinct IL6-Mediated Therapy Resistance in Pancreatic Cancer
- PMID: 35348629
- PMCID: PMC9399904
- DOI: 10.1158/2159-8290.CD-20-1484
Identification of Functional Heterogeneity of Carcinoma-Associated Fibroblasts with Distinct IL6-Mediated Therapy Resistance in Pancreatic Cancer
Abstract
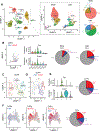
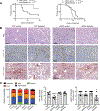
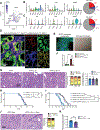
The tumor microenvironment in pancreatic ductal adenocarcinoma (PDAC) involves a significant accumulation of fibroblasts as part of the host response to cancer. Using single-cell RNA sequencing, multiplex immunostaining, and several genetic mouse models, we identify carcinoma-associated fibroblasts (CAF) with opposing functions in PDAC progression. Depletion of fibroblast activation protein (FAP)+ CAFs results in increased survival, in contrast to depletion of alpha smooth muscle actin (αSMA)+ CAFs, which leads to decreased survival. Tumor-promoting FAP+ CAFs (TP-CAF) and tumor-restraining αSMA+ CAFs (TR-CAF) differentially regulate cancer-associated pathways and accumulation of regulatory T cells. Improved efficacy of gemcitabine is observed when IL6 is deleted from αSMA+ CAFs but not from FAP+ CAFs using dual-recombinase genetic PDAC models. Improved gemcitabine efficacy due to lack of IL6 synergizes with anti-PD-1 immunotherapy to significantly improve survival of PDAC mice. Our study identifies functional heterogeneity of CAFs in PDAC progression and their different roles in therapy response.
Significance: PDAC is associated with accumulation of dense stroma consisting of fibroblasts and extracellular matrix that regulate tumor progression. Here, we identify two distinct populations of fibroblasts with opposing roles in the progression and immune landscape of PDAC. Our findings demonstrate that fibroblasts are functionally diverse with therapeutic implications. This article is highlighted in the In This Issue feature, p. 1397.
©2022 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest: KMM and YC received speaker honorarium from Stellanova Therapeutics. VSL is a Scientific Advisory Board member and stockholder of Stellanova Therapeutics.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

