Diseases 2.0: a weekly updated database of disease-gene associations from text mining and data integration
- PMID: 35348648
- PMCID: PMC9216524
- DOI: 10.1093/database/baac019
Diseases 2.0: a weekly updated database of disease-gene associations from text mining and data integration
Abstract
The scientific knowledge about which genes are involved in which diseases grows rapidly, which makes it difficult to keep up with new publications and genetics datasets. The DISEASES database aims to provide a comprehensive overview by systematically integrating and assigning confidence scores to evidence for disease-gene associations from curated databases, genome-wide association studies (GWAS) and automatic text mining of the biomedical literature. Here, we present a major update to this resource, which greatly increases the number of associations from all these sources. This is especially true for the text-mined associations, which have increased by at least 9-fold at all confidence cutoffs. We show that this dramatic increase is primarily due to adding full-text articles to the text corpus, secondarily due to improvements to both the disease and gene dictionaries used for named entity recognition, and only to a very small extent due to the growth in number of PubMed abstracts. DISEASES now also makes use of a new GWAS database, Target Illumination by GWAS Analytics, which considerably increased the number of GWAS-derived disease-gene associations. DISEASES itself is also integrated into several other databases and resources, including GeneCards/MalaCards, Pharos/Target Central Resource Database and the Cytoscape stringApp. All data in DISEASES are updated on a weekly basis and is available via a web interface at https://diseases.jensenlab.org, from where it can also be downloaded under open licenses. Database URL: https://diseases.jensenlab.org.
© The Author(s) 2022. Published by Oxford University Press.
Figures
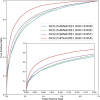
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

