Impact of defacing on automated brain atrophy estimation
- PMID: 35348936
- PMCID: PMC8964867
- DOI: 10.1186/s13244-022-01195-7
Impact of defacing on automated brain atrophy estimation
Abstract
Background: Defacing has become mandatory for anonymization of brain MRI scans; however, concerns regarding data integrity were raised. Thus, we systematically evaluated the effect of different defacing procedures on automated brain atrophy estimation.
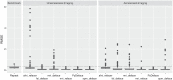
Methods: In total, 268 Alzheimer's disease patients were included from ADNI, which included unaccelerated (n = 154), within-session unaccelerated repeat (n = 67) and accelerated 3D T1 imaging (n = 114). Atrophy maps were computed using the open-source software veganbagel for every original, unmodified scan and after defacing using afni_refacer, fsl_deface, mri_deface, mri_reface, PyDeface or spm_deface, and the root-mean-square error (RMSE) between z-scores was calculated. RMSE values derived from unaccelerated and unaccelerated repeat imaging served as a benchmark. Outliers were defined as RMSE > 75th percentile and by using Grubbs's test.
Results: Benchmark RMSE was 0.28 ± 0.1 (range 0.12-0.58, 75th percentile 0.33). Outliers were found for unaccelerated and accelerated T1 imaging using the 75th percentile cutoff: afni_refacer (unaccelerated: 18, accelerated: 16), fsl_deface (unaccelerated: 4, accelerated: 18), mri_deface (unaccelerated: 0, accelerated: 15), mri_reface (unaccelerated: 0, accelerated: 2) and spm_deface (unaccelerated: 0, accelerated: 7). PyDeface performed best with no outliers (unaccelerated mean RMSE 0.08 ± 0.05, accelerated mean RMSE 0.07 ± 0.05). The following outliers were found according to Grubbs's test: afni_refacer (unaccelerated: 16, accelerated: 13), fsl_deface (unaccelerated: 10, accelerated: 21), mri_deface (unaccelerated: 7, accelerated: 20), mri_reface (unaccelerated: 7, accelerated: 6), PyDeface (unaccelerated: 5, accelerated: 8) and spm_deface (unaccelerated: 10, accelerated: 12).
Conclusion: Most defacing approaches have an impact on atrophy estimation, especially in accelerated 3D T1 imaging. Only PyDeface showed good results with negligible impact on atrophy estimation.
Keywords: Atrophy; Brain; De-identification; Magnetic resonance imaging; Privacy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Emeršič Ž, Štruc V, Peer P. Ear recognition: more than a survey. Neurocomputing. 2017;255:26–39. doi: 10.1016/j.neucom.2016.08.139. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

