Crystal structure and metal binding properties of the periplasmic iron component EfeM from Pseudomonas syringae EfeUOB/M iron-transport system
- PMID: 35348940
- PMCID: PMC9174327
- DOI: 10.1007/s10534-022-00389-2
Crystal structure and metal binding properties of the periplasmic iron component EfeM from Pseudomonas syringae EfeUOB/M iron-transport system
Abstract
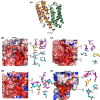
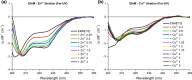
EfeUOB/M has been characterised in Pseudomonas syringae pathovar. syringae as a novel type of ferrous-iron transporter, consisting of an inner-membrane protein (EfeUPsy) and three periplasmic proteins (EfeOPsy, EfeMPsy and EfeBPsy). The role of an iron permease and peroxidase function has been identified for the EfeU and EfeB proteins, respectively, but the role of EfeO/M remains unclear. EfeMPsy is an 'M75-only' EfeO-like protein with a C-terminal peptidase-M75 domain (EfeOII/EfeM family). Herein, we report the 1.6 Å resolution crystal structure of EfeMPsy, the first structural report for an EfeM component of P. syringae pv. syringae. The structure possesses the bi-lobate architecture found in other bacterial periplasmic substrate/solute binding proteins. Metal binding studies, using SRCD and ICP-OES, reveal a preference of EfeMPsy for copper, iron and zinc. This work provides detailed knowledge of the structural scaffold, the metal site geometry, and the divalent metal binding potential of EfeM. This work provides crucial underpinning for a more detailed understanding of the role of EfeM/EfeO proteins and the peptidase-M75 domains in EfeUOB/M iron uptake systems in bacteria.
Keywords: Acidic patch; EfeM; EfeUOB iron-transport system; Metal-binding; Peptidase-M75 domain; X-ray crystallography.
© 2022. The Author(s).
Conflict of interest statement
All authors have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

