Bats host the most virulent-but not the most dangerous-zoonotic viruses
- PMID: 35349342
- PMCID: PMC9168486
- DOI: 10.1073/pnas.2113628119
Bats host the most virulent-but not the most dangerous-zoonotic viruses
Abstract
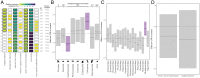
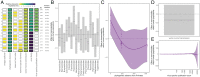
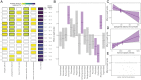
SignificanceThe clear need to mitigate zoonotic risk has fueled increased viral discovery in specific reservoir host taxa. We show that a combination of viral and reservoir traits can predict zoonotic virus virulence and transmissibility in humans, supporting the hypothesis that bats harbor exceptionally virulent zoonoses. However, pandemic prevention requires thinking beyond zoonotic capacity, virulence, and transmissibility to consider collective "burden" on human health. For this, viral discovery targeting specific reservoirs may be inefficient as death burden correlates with viral, not reservoir, traits, and depends on context-specific epidemiological dynamics across and beyond the human-animal interface. These findings suggest that longitudinal studies of viral dynamics in reservoir and spillover host populations may offer the most effective strategy for mitigating zoonotic risk.
Keywords: bat viruses; death burden; emerging infectious diseases; virulence; zoonotic viruses.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

