Studies of a mosaic patient with DBA and chimeric mice reveal erythroid cell-extrinsic contributions to erythropoiesis
- PMID: 35349664
- PMCID: PMC9185154
- DOI: 10.1182/blood.2021013507
Studies of a mosaic patient with DBA and chimeric mice reveal erythroid cell-extrinsic contributions to erythropoiesis
Abstract
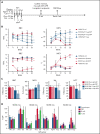
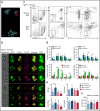
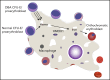
We follow a patient with Diamond-Blackfan anemia (DBA) mosaic for a pathogenic RPS19 haploinsufficiency mutation with persistent transfusion-dependent anemia. Her anemia remitted on eltrombopag (EPAG), but surprisingly, mosaicism was unchanged, suggesting that both mutant and normal cells responded. When EPAG was withheld, her anemia returned. In addition to expanding hematopoietic stem/progenitor cells, EPAG aggressively chelates iron. Because DBA anemia, at least in part, results from excessive intracellular heme leading to ferroptotic cell death, we hypothesized that the excess heme accumulating in ribosomal protein-deficient erythroid precursors inhibited the growth of adjacent genetically normal precursors, and that the efficacy of EPAG reflected its ability to chelate iron, limit heme synthesis, and thus limit toxicity in both mutant and normal cells. To test this, we studied Rpl11 haploinsufficient (DBA) mice and mice chimeric for the cytoplasmic heme export protein, FLVCR. Flvcr1-deleted mice have severe anemia, resembling DBA. Mice transplanted with ratios of DBA to wild-type marrow cells of 50:50 are anemic, like our DBA patient. In contrast, mice transplanted with Flvcr1-deleted (unable to export heme) and wild-type marrow cells at ratios of 50:50 or 80:20 have normal numbers of red cells. Additional studies suggest that heme exported from DBA erythroid cells might impede the nurse cell function of central macrophages of erythroblastic islands to impair the maturation of genetically normal coadherent erythroid cells. These findings have implications for the gene therapy of DBA and may provide insights into why del(5q) myelodysplastic syndrome patients are anemic despite being mosaic for chromosome 5q deletion and loss of RPS14.
© 2022 by The American Society of Hematology.
Figures



Comment in
-
Defending the island against excess heme.Blood. 2022 Jun 9;139(23):3359-3360. doi: 10.1182/blood.2022016341. Blood. 2022. PMID: 35679077 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

