RNAspider: a webserver to analyze entanglements in RNA 3D structures
- PMID: 35349710
- PMCID: PMC9252836
- DOI: 10.1093/nar/gkac218
RNAspider: a webserver to analyze entanglements in RNA 3D structures
Abstract
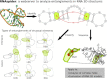
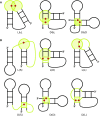
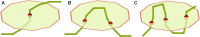
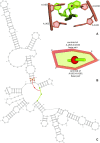
Advances in experimental and computational techniques enable the exploration of large and complex RNA 3D structures. These, in turn, reveal previously unstudied properties and motifs not characteristic for small molecules with simple architectures. Examples include entanglements of structural elements in RNA molecules and knot-like folds discovered, among others, in the genomes of RNA viruses. Recently, we presented the first classification of entanglements, determined by their topology and the type of entangled structural elements. Here, we introduce RNAspider - a web server to automatically identify, classify, and visualize primary and higher-order entanglements in RNA tertiary structures. The program applies to evaluate RNA 3D models obtained experimentally or by computational prediction. It supports the analysis of uncommon topologies in the pseudoknotted RNA structures. RNAspider is implemented as a publicly available tool with a user-friendly interface and can be freely accessed at https://rnaspider.cs.put.poznan.pl/.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Giegé R., Jühling F., Pütz J., Stadler P., Sauter C., Florentz C.. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip. Rev. RNA. 2011; 3:37–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

