Roles of RNA Modifications in Diverse Cellular Functions
- PMID: 35350378
- PMCID: PMC8957929
- DOI: 10.3389/fcell.2022.828683
Roles of RNA Modifications in Diverse Cellular Functions
Abstract
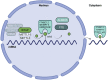
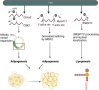
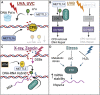
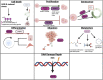
Chemical modifications of RNA molecules regulate both RNA metabolism and fate. The deposition and function of these modifications are mediated by the actions of writer, reader, and eraser proteins. At the cellular level, RNA modifications regulate several cellular processes including cell death, proliferation, senescence, differentiation, migration, metabolism, autophagy, the DNA damage response, and liquid-liquid phase separation. Emerging evidence demonstrates that RNA modifications play active roles in the physiology and etiology of multiple diseases due to their pervasive roles in cellular functions. Here, we will summarize recent advances in the regulatory and functional role of RNA modifications in these cellular functions, emphasizing the context-specific roles of RNA modifications in mammalian systems. As m6A is the best studied RNA modification in biological processes, this review will summarize the emerging advances on the diverse roles of m6A in cellular functions. In addition, we will also provide an overview for the cellular functions of other RNA modifications, including m5C and m1A. Furthermore, we will also discuss the roles of RNA modifications within the context of disease etiologies and highlight recent advances in the development of therapeutics that target RNA modifications. Elucidating these context-specific functions will increase our understanding of how these modifications become dysregulated during disease pathogenesis and may provide new opportunities for improving disease prevention and therapy by targeting these pathways.
Keywords: cellular functions; epitranscriptomics; m1A; m5C; m6A; therapeutics.
Copyright © 2022 Wilkinson, Cui and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

