Contribution of CACNA1H Variants in Autism Spectrum Disorder Susceptibility
- PMID: 35350424
- PMCID: PMC8957782
- DOI: 10.3389/fpsyt.2022.858238
Contribution of CACNA1H Variants in Autism Spectrum Disorder Susceptibility
Abstract
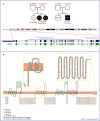
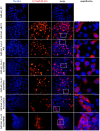
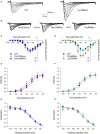
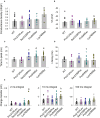
Autism Spectrum Disorder (ASD) is a highly heterogeneous neuropsychiatric disorder with a strong genetic component. The genetic architecture is complex, consisting of a combination of common low-risk and more penetrant rare variants. Voltage-gated calcium channels (VGCCs or Cav) genes have been implicated as high-confidence susceptibility genes for ASD, in accordance with the relevant role of calcium signaling in neuronal function. In order to further investigate the involvement of VGCCs rare variants in ASD susceptibility, we performed whole genome sequencing analysis in a cohort of 105 families, composed of 124 ASD individuals, 210 parents and 58 unaffected siblings. We identified 53 rare inherited damaging variants in Cav genes, including genes coding for the principal subunit and genes coding for the auxiliary subunits, in 40 ASD families. Interestingly, biallelic rare damaging missense variants were detected in the CACNA1H gene, coding for the T-type Cav3.2 channel, in ASD probands from two different families. Thus, to clarify the role of these CACNA1H variants on calcium channel activity we performed electrophysiological analysis using whole-cell patch clamp technology. Three out of four tested variants were shown to mildly affect Cav3.2 channel current density and activation properties, possibly leading to a dysregulation of intracellular Ca2+ ions homeostasis, thus altering calcium-dependent neuronal processes and contributing to ASD etiology in these families. Our results provide further support for the role of CACNA1H in neurodevelopmental disorders and suggest that rare CACNA1H variants may be involved in ASD development, providing a high-risk genetic background.
Keywords: ASD; CACNA1H; Cav3.2; VGCCs; calcium channel; rare variants.
Copyright © 2022 Viggiano, D'Andrea, Cameli, Posar, Visconti, Scaduto, Colucci, Rochat, Ceroni, Milazzo, Fucile, Maestrini and Bacchelli.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Maenner MJ, Shaw KA, Baio J, Washington A, Patrick M, DiRienzo M, et al. . Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 Sites, United States, 2016. MMWR Surveill Summ. (2020) 69:1–12. 10.15585/mmwr.ss6904a1 - DOI - PMC - PubMed
-
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders, 5th Edn. Washington, DC: American Psychiatric Association; (2013). 10.1176/appi.books.9780890425596 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

