The Oncogenic Effects, Pathways, and Target Molecules of JC Polyoma Virus T Antigen in Cancer Cells
- PMID: 35350574
- PMCID: PMC8958009
- DOI: 10.3389/fonc.2022.744886
The Oncogenic Effects, Pathways, and Target Molecules of JC Polyoma Virus T Antigen in Cancer Cells
Abstract
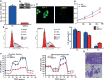
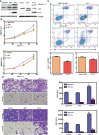
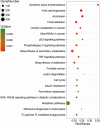
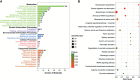
JC polyoma virus (JCPyV) is a ubiquitous polyoma virus that infects the individual to cause progressive multifocal leukoencephalopathy and malignancies. Here, we found that T-antigen knockdown suppressed proliferation, glycolysis, mitochondrial respiration, migration, and invasion, and induced apoptosis and G2 arrest. The reverse was true for T-antigen overexpression, with overexpression of Akt, survivin, retinoblastoma protein, β-catenin, β-transducin repeat-containing protein (TRCP), and inhibitor of growth (ING)1, and the underexpression of mammalian target of rapamycin (mTOR), phosphorylated (p)-mTOR, p-p38, Cyclin D1, p21, vascular endothelial growth factor (VEGF), ING2, and ING4 in hepatocellular and pancreatic cancer cells and tissues. In lens tumor cells, T antigen transcriptionally targeted viral carcinogenesis, microRNAs in cancer, focal adhesion, p53, VEGF, phosphoinositide 3 kinase-Akt, and Forkhead box O signaling pathways, fructose and mannose metabolism, ribosome biosynthesis, and choline and pyrimidine metabolism. At a metabolomics level, it targeted protein digestion and absorption, aminoacryl-tRNA biosynthesis, biosynthesis of amino acids, and the AMPK signal pathway. At a proteomic level, it targeted ribosome biogenesis in eukaryotes, citrate cycle, carbon metabolism, protein digestion and absorption, aminoacryl-tRNA biosynthesis, extracellular-matrix-receptor interaction, and biosynthesis of amino acids. In lens tumor cells, T antigen might interact with various keratins, ribosomal proteins, apolipoproteins, G proteins, ubiquitin-related proteins, RPL19, β-catenin, β-TRCP, p53, and CCAAT-enhancer-binding proteins in lens tumor cells. T antigen induced a more aggressive phenotype in mouse and human cancer cells due to oncogene activation, inactivation of tumor suppressors, and disruption of metabolism, cell adhesion, and long noncoding RNA-microRNA-target axes.
Keywords: JC virus T antigen; PI3k-Akt signal pathway; WNT/beta-catenin pathway; oncogenesis; signal pathway.
Copyright © 2022 Zheng, Xue, Jin, Jiang and Cui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The oncogenic roles of JC polyomavirus in cancer.Front Oncol. 2022 Sep 23;12:976577. doi: 10.3389/fonc.2022.976577. eCollection 2022. Front Oncol. 2022. PMID: 36212474 Free PMC article. Review.
-
The oncogenic role of JC virus T antigen in lens tumors without cell specificity of alternative splicing of its intron.Oncotarget. 2015 Apr 10;6(10):8036-45. doi: 10.18632/oncotarget.3507. Oncotarget. 2015. PMID: 25868857 Free PMC article.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
-
Loss of G3BP1 suppresses proliferation, migration, and invasion of esophageal cancer cells via Wnt/β-catenin and PI3K/AKT signaling pathways.J Cell Physiol. 2019 Nov;234(11):20469-20484. doi: 10.1002/jcp.28648. Epub 2019 Apr 15. J Cell Physiol. 2019. PMID: 30989663
-
New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia.Cell Signal. 2014 Jan;26(1):149-61. doi: 10.1016/j.cellsig.2013.09.021. Epub 2013 Oct 16. Cell Signal. 2014. PMID: 24140475 Review.
Cited by
-
The oncogenic roles of JC polyomavirus in cancer.Front Oncol. 2022 Sep 23;12:976577. doi: 10.3389/fonc.2022.976577. eCollection 2022. Front Oncol. 2022. PMID: 36212474 Free PMC article. Review.
-
The Effects of Deregulated Ribosomal Biogenesis in Cancer.Biomolecules. 2023 Oct 30;13(11):1593. doi: 10.3390/biom13111593. Biomolecules. 2023. PMID: 38002277 Free PMC article. Review.
References
-
- Meinke G, Phelan PJ, Shin J, Gagnon D, Archambault J, Bohm A, et al. . Structural Based Analyses of the JC Virus T-Antigen F258L Mutant Provides Evidence for DNA Dependent Conformational Changes in the C-Termini of Polyoma Virus Origin Binding Domains. PloS Pathog (2016) 12(1):e1005362. doi: 10.1371/journal.ppat.1005362 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

