Applications of Coarse-Grained Models in Metabolic Engineering
- PMID: 35350716
- PMCID: PMC8957914
- DOI: 10.3389/fmolb.2022.806213
Applications of Coarse-Grained Models in Metabolic Engineering
Abstract
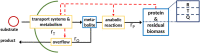
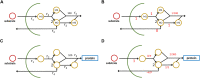
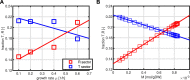
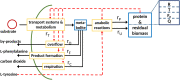
Mathematical modeling is a promising tool for better understanding of cellular processes. In recent years, the development of coarse-grained models has gained attraction since these simple models are able to capture and describe a broad range of growth conditions. Coarse-grained models often comprise only two cellular components, a low molecular component as representative for central metabolism and energy generation and a macromolecular component, representing the entire proteome. A framework is presented that presents a strict mass conservative model for bacterial growth during a biotechnological production process. After providing interesting properties for the steady-state solution, applications are presented 1) for a production process of an amino acid and 2) production of a metabolite from central metabolism.
Keywords: kinetic model; mathematical modeling; metabolic and gene regulatory networks; metabolic engineering; microbial growth; resource allocation; systems biotechnology.
Copyright © 2022 Doan, Hoang, Heins and Kremling.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




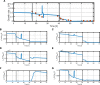
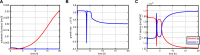
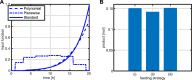
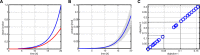
References
LinkOut - more resources
Full Text Sources

