A primer to traction force microscopy
- PMID: 35351517
- PMCID: PMC9092999
- DOI: 10.1016/j.jbc.2022.101867
A primer to traction force microscopy
Abstract
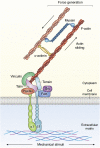
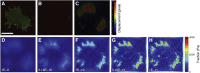
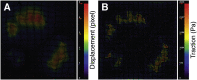
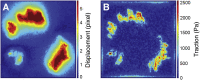
Traction force microscopy (TFM) has emerged as a versatile technique for the measurement of single-cell-generated forces. TFM has gained wide use among mechanobiology laboratories, and several variants of the original methodology have been proposed. However, issues related to the experimental setup and, most importantly, data analysis of cell traction datasets may restrain the adoption of TFM by a wider community. In this review, we summarize the state of the art in TFM-related research, with a focus on the analytical methods underlying data analysis. We aim to provide the reader with a friendly compendium underlying the potential of TFM and emphasizing the methodological framework required for a thorough understanding of experimental data. We also compile a list of data analytics tools freely available to the scientific community for the furtherance of knowledge on this powerful technique.
Keywords: biophysics; cell adhesion; cytoskeleton; focal adhesion; mechanosignaling; mechanotransduction; traction force microscopy.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

