Global transcriptomic characterization of T cells in individuals with chronic HIV-1 infection
- PMID: 35351857
- PMCID: PMC8964811
- DOI: 10.1038/s41421-021-00367-x
Global transcriptomic characterization of T cells in individuals with chronic HIV-1 infection
Abstract
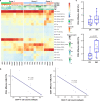
To obtain a comprehensive scenario of T cell profiles and synergistic immune responses, we performed single-cell RNA sequencing (scRNA-seq) on the peripheral T cells of 14 individuals with chronic human immunodeficiency virus 1 (HIV-1) infection, including nine treatment-naive (TP) and eight antiretroviral therapy (ART) participants (of whom three were paired with TP cases), and compared the results with four healthy donors (HD). Through analyzing the transcriptional profiles of CD4+ and CD8+ T cells, coupled with assembled T cell receptor sequences, we observed the significant loss of naive T cells, prolonged inflammation, and increased response to interferon-α in TP individuals, which could be partially restored by ART. Interestingly, we revealed that CD4+ and CD8+ Effector-GNLY clusters were expanded in TP cases, and persistently increased in ART individuals where they were typically correlated with poor immune restoration. This transcriptional dataset enables a deeper understanding of the pathogenesis of HIV-1 infection and is also a rich resource for developing novel immune targeted therapeutic strategies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014;384:258–271. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

