Modulating DNA Repair Pathways to Diversify Genomic Alterations in Saccharomyces cerevisiae
- PMID: 35352941
- PMCID: PMC9045378
- DOI: 10.1128/spectrum.02326-21
Modulating DNA Repair Pathways to Diversify Genomic Alterations in Saccharomyces cerevisiae
Abstract
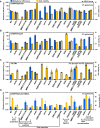
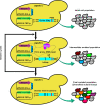
Nuclease based genome editing systems have emerged as powerful tools to drive genomic alterations and enhance genome evolution via precise engineering in the various human and microbial cells. However, error-prone DNA repair has not been well studied previously to generate diverse genomic alterations and novel phenotypes. Here, we systematically investigated the potential interplay between DNA double strand break (DSB) repair and genome editing tools, and found that modulating the DSB end resection proteins could significantly improve mutational efficiency and diversity without exogenous DNA template in yeast. Deleting SAE2, EXO1, or FUN30, or overexpressing MRE11-H125N (nuclease-dead allele of MRE11), for DSB end resection markedly increased the efficiency of CRISPR/SpCas9 (more than 22-fold) and CRISPR/AsCpf1 (more than 30-fold)-induced mutagenesis. Deleting SAE2 or overexpressing MRE11-H125N substantially diversified CRISPR/SpCas9 or AsCpf1-induced mutation 2-3-fold at URA3 locus, and 3-5-fold at ADE2 locus. Thus, the error-prone DNA repair protein was employed to develop a novel mutagenic genome editing (mGE) strategy, which can increase the mutation numbers and effectively improve the ethanol/glycerol ratio of Saccharomyces cerevisiae through modulating the expression of FPS1 and GPD1. This study highlighted the feasibility of potentially reshaping the capability of genome editing by regulating the different DSB repair proteins and can thus expand the application of genome editing in diversifying gene expression and enhancing genome evolution. IMPORTANCE Most of the published papers about nuclease-assisted genome editing focused on precision engineering in human cells. However, the topic of inducing mutagenesis via error-prone repair has often been ignored in yeast. In this study, we reported that perturbing DNA repair, especially modifications of the various DSB end resection-related proteins, could greatly improve the mutational efficiency and diversity, and thus functionally reshape the capability of the different genome editing tools without requiring an exogenous DNA template in yeast. Specifically, mutagenic genome editing (mGE) was developed based on CRISPR/AsCpf1 and MRE11-H125N overexpression, and used to generate promoters of different strengths more efficiently. Thus, this work provides a novel method to diversify gene expression and enhance genome evolution.
Keywords: DSB repair; diversified mutation; gene expression; genome editing; mutational efficiency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

