β-Lactam Resistance in Azospirillum baldaniorum Sp245 Is Mediated by Lytic Transglycosylase and β-Lactamase and Regulated by a Cascade of RpoE7→RpoH3 Sigma Factors
- PMID: 35352964
- PMCID: PMC9017315
- DOI: 10.1128/jb.00010-22
β-Lactam Resistance in Azospirillum baldaniorum Sp245 Is Mediated by Lytic Transglycosylase and β-Lactamase and Regulated by a Cascade of RpoE7→RpoH3 Sigma Factors
Abstract
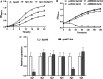
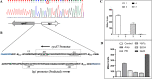
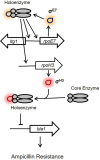
Bacterial resistance to β-lactam antibiotics is often mediated by β-lactamases and lytic transglycosylases. Azospirillum baldaniorum Sp245 is a plant-growth-promoting rhizobacterium that shows high levels of resistance to ampicillin. Investigating the molecular basis of ampicillin resistance and its regulation in A. baldaniorum Sp245, we found that a gene encoding lytic transglycosylase (Ltg1) is organized divergently from a gene encoding an extracytoplasmic function (ECF) σ factor (RpoE7) in its genome. Inactivation of rpoE7 in A. baldaniorum Sp245 led to increased ability to form cell-cell aggregates and produce exopolysaccharides and biofilm, suggesting that rpoE7 might contribute to antibiotic resistance. Inactivation of ltg1 in A. baldaniorum Sp245, however, adversely affected its growth, indicating a requirement of Ltg1 for optimal growth. The expression of rpoE7, as well that of as ltg1, was positively regulated by RpoE7, and overexpression of RpoE7 conferred ampicillin sensitivity to both the rpoE7::km mutant and its parent. In addition, RpoE7 negatively regulated the expression of a gene encoding a β-lactamase (bla1). Out of the 5 paralogs of RpoH encoded in the genome of A. baldaniorum Sp245, RpoH3 played major roles in conferring ampicillin sensitivity and in the downregulation of bla1. The expression of rpoH3 was positively regulated by RpoE7. Collectively, these observations reveal a novel regulatory cascade of RpoE7-RpoH3 σ factors that negatively regulates ampicillin resistance in A. baldaniorum Sp245 by controlling the expression of a β-lactamase and a lytic transglycosylase. In the absence of a cognate anti-sigma factor, addressing how the activity of RpoE7 is regulated by β-lactams will unravel new mechanisms of regulation of β-lactam resistance in bacteria. IMPORTANCE Antimicrobial resistance is a global health problem that requires a better understanding of the mechanisms that bacteria use to resist antibiotics. Bacteria inhabiting the plant rhizosphere are a potential source of antibiotic resistance, but their mechanisms controlling antibiotic resistance are poorly understood. A. baldaniorum Sp245 is a rhizobacterium that is known for its characteristic resistance to ampicillin. Here, we show that an AmpC-type β-lactamase and a lytic transglycosylase mediate resistance to ampicillin in A. baldaniorum Sp245. While the gene encoding lytic transglycosylase is positively regulated by an ECF σ-factor (RpoE7), a cascade of RpoE7 and RpoH3 σ factors negatively regulates the expression of β-lactamase. This is the first evidence showing involvement of a regulatory cascade of σ factors in the regulation of ampicillin resistance in a rhizobacterium.
Keywords: ECF σ factor; RpoE7; RpoH; antibiotic resistance; bacteria; lytic transglycosylase; transcription regulation; β-lactamase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

