Emergent dynamics of a three-node regulatory network explain phenotypic switching and heterogeneity: a case study of Th1/Th2/Th17 cell differentiation
- PMID: 35353012
- PMCID: PMC9265159
- DOI: 10.1091/mbc.E21-10-0521
Emergent dynamics of a three-node regulatory network explain phenotypic switching and heterogeneity: a case study of Th1/Th2/Th17 cell differentiation
Abstract
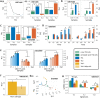
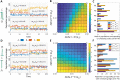
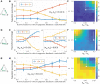
Naïve helper (CD4+) T-cells can differentiate into distinct functional subsets including Th1, Th2, and Th17 phenotypes. Each of these phenotypes has a "master regulator"-T-bet (Th1), GATA3 (Th2), and RORγT (Th17)-that inhibits the other two master regulators. Such mutual repression among them at a transcriptional level can enable multistability, giving rise to six experimentally observed phenotype, Th1, Th2, Th17, hybrid Th/Th2, hybrid Th2/Th17, and hybrid Th1/Th17. However, the dynamics of switching among these phenotypes, particularly in the case of epigenetic influence, remain unclear. Here through mathematical modeling, we investigated the coupled transcription-epigenetic dynamics in a three-node mutually repressing network to elucidate how epigenetic changes mediated by any master regulator can influence the transition rates among different cellular phenotypes. We show that the degree of plasticity exhibited by one phenotype depends on relative strength and duration of mutual epigenetic repression mediated among the master regulators in a three-node network. Further, our model predictions can offer putative mechanisms underlying relatively higher plasticity of Th17 phenotype as observed in vitro and in vivo. Together, our modeling framework characterizes phenotypic plasticity and heterogeneity as an outcome of emergent dynamics of a three-node regulatory network, such as the one mediated by T-bet/GATA3/RORγT.
Figures




Similar articles
-
Multistability and predominant hybrid phenotypes in a four node mutually repressive network of Th1/Th2/Th17/Treg differentiation.NPJ Syst Biol Appl. 2024 Oct 24;10(1):123. doi: 10.1038/s41540-024-00433-6. NPJ Syst Biol Appl. 2024. PMID: 39448615 Free PMC article.
-
Shenghua Decoction reduces uterine bleeding and regulates T-cell paradigm in human deciduas of RU486 medical abortion.J Ethnopharmacol. 2013 Dec 12;150(3):907-17. doi: 10.1016/j.jep.2013.09.033. Epub 2013 Oct 17. J Ethnopharmacol. 2013. PMID: 24140602 Clinical Trial.
-
A cellular and molecular view of T helper 17 cell plasticity in autoimmunity.J Autoimmun. 2018 Feb;87:1-15. doi: 10.1016/j.jaut.2017.12.007. Epub 2017 Dec 22. J Autoimmun. 2018. PMID: 29275836 Review.
-
Th2-biased GATA-3 transgenic mice developed severe experimental peritoneal fibrosis compared with Th1-biased T-bet and Th17-biased RORγt transgenic mice.Exp Anim. 2015;64(4):353-62. doi: 10.1538/expanim.15-0019. Epub 2015 Jul 6. Exp Anim. 2015. PMID: 26156402 Free PMC article.
-
The Mechanisms of Effector Th Cell Responses Contribute to Treg Cell Function: New Insights into Pathogenesis and Therapy of Asthma.Front Immunol. 2022 Jul 11;13:862866. doi: 10.3389/fimmu.2022.862866. eCollection 2022. Front Immunol. 2022. PMID: 35898499 Free PMC article. Review.
Cited by
-
Multistability and predominant hybrid phenotypes in a four node mutually repressive network of Th1/Th2/Th17/Treg differentiation.NPJ Syst Biol Appl. 2024 Oct 24;10(1):123. doi: 10.1038/s41540-024-00433-6. NPJ Syst Biol Appl. 2024. PMID: 39448615 Free PMC article.
-
T-helper cells flexibility: the possibility of reprogramming T cells fate.Front Immunol. 2023 Nov 1;14:1284178. doi: 10.3389/fimmu.2023.1284178. eCollection 2023. Front Immunol. 2023. PMID: 38022605 Free PMC article. Review.
-
Global quantitative understanding of non-equilibrium cell fate decision-making in response to pheromone.iScience. 2023 Sep 9;26(10):107885. doi: 10.1016/j.isci.2023.107885. eCollection 2023 Oct 20. iScience. 2023. PMID: 37766979 Free PMC article.
-
Emergent dynamics of cellular decision making in multi-node mutually repressive regulatory networks.J R Soc Interface. 2025 Aug;22(229):20250190. doi: 10.1098/rsif.2025.0190. Epub 2025 Aug 20. J R Soc Interface. 2025. PMID: 40829640 Free PMC article.
-
Network topology and interaction logic determine states it supports.NPJ Syst Biol Appl. 2024 Aug 28;10(1):98. doi: 10.1038/s41540-024-00423-8. NPJ Syst Biol Appl. 2024. PMID: 39198512 Free PMC article. Review.
References
-
- Äijö T, Edelman SM, Lönnberg T, Larjo A, Kallionpää H, Tuomela S, Engström E, Lahesmaa R, Lähdesmäki H (2012). An integrative computational systems biology approach identifies differentially regulated dynamic transcriptome signatures which drive the initiation of human T helper cell differentiation. BMC Genomics 13, 572. - PMC - PubMed
-
- Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, Fernandez B, Kelderman S, Schumacher TN, Corti D, et al. (2015). T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science 347, 400–406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

