Inflammation and Brain Structure in Schizophrenia and Other Neuropsychiatric Disorders: A Mendelian Randomization Study
- PMID: 35353173
- PMCID: PMC8968718
- DOI: 10.1001/jamapsychiatry.2022.0407
Inflammation and Brain Structure in Schizophrenia and Other Neuropsychiatric Disorders: A Mendelian Randomization Study
Abstract
Importance: Previous in vitro and postmortem research suggests that inflammation may lead to structural brain changes via activation of microglia and/or astrocytic dysfunction in a range of neuropsychiatric disorders.
Objective: To investigate the relationship between inflammation and changes in brain structures in vivo and to explore a transcriptome-driven functional basis with relevance to mental illness.
Design, setting, and participants: This study used multistage linked analyses, including mendelian randomization (MR), gene expression correlation, and connectivity analyses. A total of 20 688 participants in the UK Biobank, which includes clinical, genomic, and neuroimaging data, and 6 postmortem brains from neurotypical individuals in the Allen Human Brain Atlas (AHBA), including RNA microarray data. Data were extracted in February 2021 and analyzed between March and October 2021.
Exposures: Genetic variants regulating levels and activity of circulating interleukin 1 (IL-1), IL-2, IL-6, C-reactive protein (CRP), and brain-derived neurotrophic factor (BDNF) were used as exposures in MR analyses.
Main outcomes and measures: Brain imaging measures, including gray matter volume (GMV) and cortical thickness (CT), were used as outcomes. Associations were considered significant at a multiple testing-corrected threshold of P < 1.1 × 10-4. Differential gene expression in AHBA data was modeled in brain regions mapped to areas significant in MR analyses; genes were tested for biological and disease overrepresentation in annotation databases and for connectivity in protein-protein interaction networks.
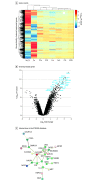
Results: Of 20 688 participants in the UK Biobank sample, 10 828 (52.3%) were female, and the mean (SD) age was 55.5 (7.5) years. In the UK Biobank sample, genetically predicted levels of IL-6 were associated with GMV in the middle temporal cortex (z score, 5.76; P = 8.39 × 10-9), inferior temporal (z score, 3.38; P = 7.20 × 10-5), fusiform (z score, 4.70; P = 2.60 × 10-7), and frontal (z score, -3.59; P = 3.30 × 10-5) cortex together with CT in the superior frontal region (z score, -5.11; P = 3.22 × 10-7). No significant associations were found for IL-1, IL-2, CRP, or BDNF after correction for multiple comparison. In the AHBA sample, 5 of 6 participants (83%) were male, and the mean (SD) age was 42.5 (13.4) years. Brain-wide coexpression analysis showed a highly interconnected network of genes preferentially expressed in the middle temporal gyrus (MTG), which further formed a highly connected protein-protein interaction network with IL-6 (enrichment test of expected vs observed network given the prevalence and degree of interactions in the STRING database: 43 nodes/30 edges observed vs 8 edges expected; mean node degree, 1.4; genome-wide significance, P = 4.54 × 10-9). MTG differentially expressed genes that were functionally enriched for biological processes in schizophrenia, autism spectrum disorder, and epilepsy.
Conclusions and relevance: In this study, genetically determined IL-6 was associated with brain structure and potentially affects areas implicated in developmental neuropsychiatric disorders, including schizophrenia and autism.
Conflict of interest statement
Figures




References
-
- Kappelmann N, Arloth J, Georgakis MK, et al. . Dissecting the association between inflammation, metabolic dysregulation, and specific depressive symptoms: a genetic correlation and 2-sample mendelian randomization study. JAMA Psychiatry. 2021;78(2):161-170. doi:10.1001/jamapsychiatry.2020.3436 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/S037675/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00002/7/MRC_/Medical Research Council/United Kingdom
- MC_PC_17213/MRC_/Medical Research Council/United Kingdom
- MR/S003991/1/MRC_/Medical Research Council/United Kingdom
- 204623/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- MR/V004905/1/MRC_/Medical Research Council/United Kingdom
- 201486/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MR/W014416/1/MRC_/Medical Research Council/United Kingdom
- MR/N029488/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

