Applied phenomics and genomics for improving barley yellow dwarf resistance in winter wheat
- PMID: 35353191
- PMCID: PMC9258586
- DOI: 10.1093/g3journal/jkac064
Applied phenomics and genomics for improving barley yellow dwarf resistance in winter wheat
Abstract
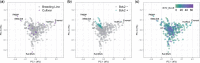
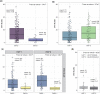
Barley yellow dwarf is one of the major viral diseases of cereals. Phenotyping barley yellow dwarf in wheat is extremely challenging due to similarities to other biotic and abiotic stresses. Breeding for resistance is additionally challenging as the wheat primary germplasm pool lacks genetic resistance, with most of the few resistance genes named to date originating from a wild relative species. The objectives of this study were to (1) evaluate the use of high-throughput phenotyping to improve barley yellow dwarf assessment; (2) identify genomic regions associated with barley yellow dwarf resistance; and (3) evaluate the ability of genomic selection models to predict barley yellow dwarf resistance. Up to 107 wheat lines were phenotyped during each of 5 field seasons under both insecticide treated and untreated plots. Across all seasons, barley yellow dwarf severity was lower within the insecticide treatment along with increased plant height and grain yield compared with untreated entries. Only 9.2% of the lines were positive for the presence of the translocated segment carrying the resistance gene Bdv2. Despite the low frequency, this region was identified through association mapping. Furthermore, we mapped a potentially novel genomic region for barley yellow dwarf resistance on chromosome 5AS. Given the variable heritability of the trait (0.211-0.806), we obtained a predictive ability for barley yellow dwarf severity ranging between 0.06 and 0.26. Including the presence or absence of Bdv2 as a covariate in the genomic selection models had a large effect for predicting barley yellow dwarf but almost no effect for other observed traits. This study was the first attempt to characterize barley yellow dwarf using field-high-throughput phenotyping and apply genomic selection to predict disease severity. These methods have the potential to improve barley yellow dwarf characterization, additionally identifying new sources of resistance will be crucial for delivering barley yellow dwarf resistant germplasm.
Keywords: Triticum aestivum; barley yellow dwarf (BYD); genome-wide association mapping (GWAS); genomic selection (GS); high-throughput phenotyping (HTP); resistance; tolerance; virus.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
-
- Ayala-Navarrete L, Larkin P.. Wheat virus diseases: breeding for resistance and tolerance. In: Bonjean A, Angus W, van Ginkel M, editors. World Wheat Book: A History of Wheat Breeding, Vol. 2. Paris: Lavoisier; 2011. p. 1073–1107.
-
- Ayala L, Henry M, van Ginkel M, Singh R, Keller B, Khairallah M.. Identification of QTLs for BYDV tolerance in bread wheat. Euphytica. 2002;128(2):249–259. doi:10.1023/A:1020883414410. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
