GROMACS in the Cloud: A Global Supercomputer to Speed Up Alchemical Drug Design
- PMID: 35353508
- PMCID: PMC9006219
- DOI: 10.1021/acs.jcim.2c00044
GROMACS in the Cloud: A Global Supercomputer to Speed Up Alchemical Drug Design
Abstract
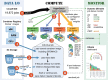
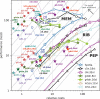
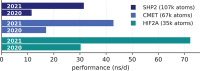
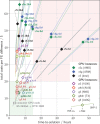
We assess costs and efficiency of state-of-the-art high-performance cloud computing and compare the results to traditional on-premises compute clusters. Our use case is atomistic simulations carried out with the GROMACS molecular dynamics (MD) toolkit with a particular focus on alchemical protein-ligand binding free energy calculations. We set up a compute cluster in the Amazon Web Services (AWS) cloud that incorporates various different instances with Intel, AMD, and ARM CPUs, some with GPU acceleration. Using representative biomolecular simulation systems, we benchmark how GROMACS performs on individual instances and across multiple instances. Thereby we assess which instances deliver the highest performance and which are the most cost-efficient ones for our use case. We find that, in terms of total costs, including hardware, personnel, room, energy, and cooling, producing MD trajectories in the cloud can be about as cost-efficient as an on-premises cluster given that optimal cloud instances are chosen. Further, we find that high-throughput ligand-screening can be accelerated dramatically by using global cloud resources. For a ligand screening study consisting of 19 872 independent simulations or ∼200 μs of combined simulation trajectory, we made use of diverse hardware available in the cloud at the time of the study. The computations scaled-up to reach peak performance using more than 4 000 instances, 140 000 cores, and 3 000 GPUs simultaneously. Our simulation ensemble finished in about 2 days in the cloud, while weeks would be required to complete the task on a typical on-premises cluster consisting of several hundred nodes.
Conflict of interest statement
The authors declare the following competing financial interest(s): Christian Kniep, Austin Cherian, and Ludvig Nordstrom are employees of Amazon.com, Inc.
Figures





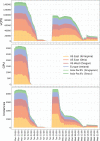
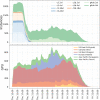
References
-
- Wang L.; Wu Y.; Deng Y.; Kim B.; Pierce L.; Krilov G.; Lupyan D.; Robinson S.; Dahlgren M. K.; Greenwood J.; Romero D. L.; Masse C.; Knight J. L.; Steinbrecher T.; Beuming T.; Damm W.; Harder E.; Sherman W.; Brewer M.; Wester R.; Murcko M.; Frye L.; Farid R.; Lin T.; Mobley D. L.; Jorgensen W. L.; Berne B. J.; Friesner R. A.; Abel R. J. Am. Chem. Soc. 2015, 137, 2695–2703. 10.1021/ja512751q. - DOI - PubMed
-
- Schindler C. E. M.; Baumann H.; Blum A.; Böse D.; Buchstaller H.-P.; Burgdorf L.; Cappel D.; Chekler E.; Czodrowski P.; Dorsch D.; Eguida M. K. I.; Follows B.; Fuchß T.; Grädler U.; Gunera J.; Johnson T.; Jorand Lebrun C.; Karra S.; Klein M.; Knehans T.; Koetzner L.; Krier M.; Leiendecker M.; Leuthner B.; Li L.; Mochalkin I.; Musil D.; Neagu C.; Rippmann F.; Schiemann K.; Schulz R.; Steinbrecher T.; Tanzer E.-M.; Unzue Lopez A.; Viacava Follis A.; Wegener A.; Kuhn D. J. Chem. Inf. Model. 2020, 60, 5457–5474. 10.1021/acs.jcim.0c00900. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

