Differential gene expression reveals host factors for viral shedding variation in mallards (Anas platyrhynchos) infected with low-pathogenic avian influenza virus
- PMID: 35353676
- PMCID: PMC10519146
- DOI: 10.1099/jgv.0.001724
Differential gene expression reveals host factors for viral shedding variation in mallards (Anas platyrhynchos) infected with low-pathogenic avian influenza virus
Abstract
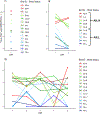
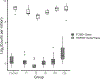
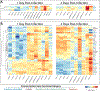
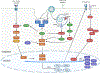
Intraspecific variation in pathogen shedding impacts disease transmission dynamics; therefore, understanding the host factors associated with individual variation in pathogen shedding is key to controlling and preventing outbreaks. In this study, ileum and bursa of Fabricius tissues of wild-bred mallards (Anas platyrhynchos) infected with low-pathogenic avian influenza (LPAIV) were evaluated at various post-infection time points to determine genetic host factors associated with intraspecific variation in viral shedding. By analysing transcriptome sequencing data (RNA-seq), we found that LPAIV-infected wild-bred mallards do not exhibit differential gene expression compared to uninfected birds, but that gene expression was associated with cloacal viral shedding quantity early in the infection. In both tissues, immune gene expression was higher in high/moderate shedding birds compared to low shedding birds, and significant positive relationships with viral shedding were observed. In the ileum, expression for host genes involved in viral cell entry was lower in low shedders compared to moderate shedders at 1 day post-infection (DPI), and expression for host genes promoting viral replication was higher in high shedders compared to low shedders at 2 DPI. Our findings indicate that viral shedding is a key factor for gene expression differences in LPAIV-infected wild-bred mallards, and the genes identified in this study could be important for understanding the molecular mechanisms driving intraspecific variation in pathogen shedding.
Keywords: influenza; shedding; transcriptomics; virus.
Figures
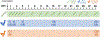


References
-
- Vanderwaal KL, Ezenwa VO. Heterogeneity in pathogen transmission: mechanisms and methodology. Funct Ecol. 2016;30:1606–22.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

