Cross-reactivity of glycan-reactive HIV-1 broadly neutralizing antibodies with parasite glycans
- PMID: 35354052
- PMCID: PMC10073069
- DOI: 10.1016/j.celrep.2022.110611
Cross-reactivity of glycan-reactive HIV-1 broadly neutralizing antibodies with parasite glycans
Abstract
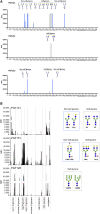
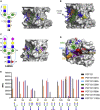
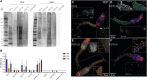
The HIV-1 Envelope glycoprotein (Env) is the sole target for broadly neutralizing antibodies (bnAbs). Env is heavily glycosylated with host-derived N-glycans, and many bnAbs bind to, or are dependent upon, Env glycans for neutralization. Although glycan-binding bnAbs are frequently detected in HIV-infected individuals, attempts to elicit them have been unsuccessful because of the poor immunogenicity of Env N-glycans. Here, we report cross-reactivity of glycan-binding bnAbs with self- and non-self N-glycans and glycoprotein antigens from different life-stages of Schistosoma mansoni. Using the IAVI Protocol C HIV infection cohort, we examine the relationship between S. mansoni seropositivity and development of bnAbs targeting glycan-dependent epitopes. We show that the unmutated common ancestor of the N332/V3-specific bnAb lineage PCDN76, isolated from an HIV-infected donor with S. mansoni seropositivity, binds to S. mansoni cercariae while lacking reactivity to gp120. Overall, these results present a strategy for elicitation of glycan-reactive bnAbs which could be exploited in HIV-1 vaccine development.
Keywords: CP: Immunology; HIV-1; Schisotosoma mansoni; glycan epitope; neutralizing antibody; vaccine.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
Shared sugars - parasite glycan homology in HIV-1 vaccine design.Trends Parasitol. 2022 Jul;38(7):498-500. doi: 10.1016/j.pt.2022.04.001. Epub 2022 Apr 29. Trends Parasitol. 2022. PMID: 35501266
References
-
- Angulo J., Nieto P.M. STD-NMR: application to transient interactions between biomolecules-a quantitative approach. Eur. Biophys. J. 2011;40:1357–1369. - PubMed
-
- De Boer A.R., Hokke C.H., Deelder A.M., Wuhrer M. General microarray technique for immobilization and screening of natural glycans. Anal. Chem. 2007;79:8107–8113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/M011216/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K024426/1/MRC_/Medical Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
- BB/P010660/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

