Pervasive Transcription-coupled DNA repair in E. coli
- PMID: 35354807
- PMCID: PMC8967931
- DOI: 10.1038/s41467-022-28871-y
Pervasive Transcription-coupled DNA repair in E. coli
Abstract
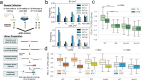
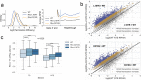
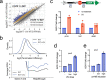
Global Genomic Repair (GGR) and Transcription-Coupled Repair (TCR) have been viewed, respectively, as major and minor sub-pathways of the nucleotide excision repair (NER) process that removes bulky lesions from the genome. Here we applied a next generation sequencing assay, CPD-seq, in E. coli to measure the levels of cyclobutane pyrimidine dimer (CPD) lesions before, during, and after UV-induced genotoxic stress, and, therefore, to determine the rate of genomic recovery by NER at a single nucleotide resolution. We find that active transcription is necessary for the repair of not only the template strand (TS), but also the non-template strand (NTS), and that the bulk of TCR is independent of Mfd - a DNA translocase that is thought to be necessary and sufficient for TCR in bacteria. We further show that repair of both TS and NTS is enhanced by increased readthrough past Rho-dependent terminators. We demonstrate that UV-induced genotoxic stress promotes global antitermination so that TCR is more accessible to the antisense, intergenic, and other low transcribed regions. Overall, our data suggest that GGR and TCR are essentially the same process required for complete repair of the bacterial genome.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. - PubMed
-
- Hanawalt, P. C. & Haynes, R. H. The repair of DNA. Scientific American216, 36–43 (1967). - PubMed
-
- Van Houten B, Gamper H, Hearst JE, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem. 1988;263:16553–16560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

