Detection and quantification of infectious severe acute respiratory coronavirus-2 in diverse clinical and environmental samples
- PMID: 35354854
- PMCID: PMC8967087
- DOI: 10.1038/s41598-022-09218-5
Detection and quantification of infectious severe acute respiratory coronavirus-2 in diverse clinical and environmental samples
Abstract
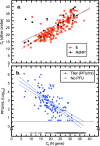
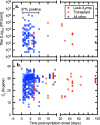
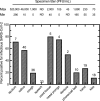
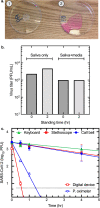
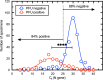
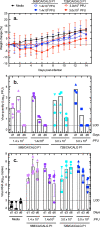
To explore the potential modes of Severe Acute Respiratory Coronavirus-2 (SARS-CoV-2) transmission, we collected 535 diverse clinical and environmental samples from 75 infected hospitalized and community patients. Infectious SARS-CoV-2 with quantitative burdens varying from 5 plaque-forming units/mL (PFU/mL) up to 1.0 × 106 PFU/mL was detected in 151/459 (33%) of the specimens assayed and up to 1.3 × 106 PFU/mL on fomites with confirmation by plaque morphology, PCR, immunohistochemistry, and/or sequencing. Infectious virus in clinical and associated environmental samples correlated with time since symptom onset with no detection after 7-8 days in immunocompetent hosts and with N-gene based Ct values ≤ 25 significantly predictive of yielding plaques in culture. SARS-CoV-2 isolated from patient respiratory tract samples caused illness in a hamster model with a minimum infectious dose of ≤ 14 PFU. Together, our findings offer compelling evidence that large respiratory droplet and contact (direct and indirect i.e., fomites) are important modes of SARS-CoV-2 transmission.
© 2022. The Author(s).
Conflict of interest statement
All authors have completed the ICMJE uniform disclosure form at
Figures



References
-
- Centers for Disease Control and Prevention. Public health activity guidance. Centers for Disease Control and Preventionhttps://www.cdc.gov/coronavirus/2019-ncov/hcp/non-covid-19-client-intera... (2020).
-
- World Health Organization. Prevention, identification and management of health worker infection in the context of COVID-19: interim guidance. WHO Document Number: WHO/2019-nCoV/HW_infection/2020 (2020).
-
- World Health Organization. Infection prevention and control of epidemic- and pandemic-prone acute respiratory infections in health care (2014). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

