Anatomic position determines oncogenic specificity in melanoma
- PMID: 35355015
- PMCID: PMC9355078
- DOI: 10.1038/s41586-022-04584-6
Anatomic position determines oncogenic specificity in melanoma
Abstract
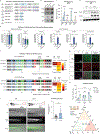
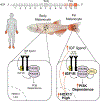
Oncogenic alterations to DNA are not transforming in all cellular contexts1,2. This may be due to pre-existing transcriptional programmes in the cell of origin. Here we define anatomic position as a major determinant of why cells respond to specific oncogenes. Cutaneous melanoma arises throughout the body, whereas the acral subtype arises on the palms of the hands, soles of the feet or under the nails3. We sequenced the DNA of cutaneous and acral melanomas from a large cohort of human patients and found a specific enrichment for BRAF mutations in cutaneous melanoma and enrichment for CRKL amplifications in acral melanoma. We modelled these changes in transgenic zebrafish models and found that CRKL-driven tumours formed predominantly in the fins of the fish. The fins are the evolutionary precursors to tetrapod limbs, indicating that melanocytes in these acral locations may be uniquely susceptible to CRKL. RNA profiling of these fin and limb melanocytes, when compared with body melanocytes, revealed a positional identity gene programme typified by posterior HOX13 genes. This positional gene programme synergized with CRKL to amplify insulin-like growth factor (IGF) signalling and drive tumours at acral sites. Abrogation of this CRKL-driven programme eliminated the anatomic specificity of acral melanoma. These data suggest that the anatomic position of the cell of origin endows it with a unique transcriptional state that makes it susceptible to only certain oncogenic insults.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures










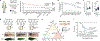



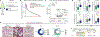

Comment in
-
Cell position matters in tumour development.Nature. 2022 Apr;604(7905):248-250. doi: 10.1038/d41586-022-00856-3. Nature. 2022. PMID: 35354976 No abstract available.
References
-
- Reed R In New Concepts in Surgical Pathology of the Skin 89–90 (Wiley, 1976).
-
- Curtin JA et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 353, 2135–2147 (2005). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

