Genetic instability from a single S phase after whole-genome duplication
- PMID: 35355016
- PMCID: PMC8986533
- DOI: 10.1038/s41586-022-04578-4
Genetic instability from a single S phase after whole-genome duplication
Erratum in
-
Author Correction: Genetic instability from a single S phase after whole-genome duplication.Nature. 2022 Aug;608(7923):E27. doi: 10.1038/s41586-022-05099-w. Nature. 2022. PMID: 35864235 Free PMC article. No abstract available.
Abstract
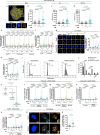
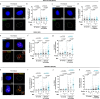
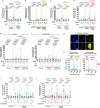
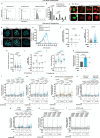
Diploid and stable karyotypes are associated with health and fitness in animals. By contrast, whole-genome duplications-doublings of the entire complement of chromosomes-are linked to genetic instability and frequently found in human cancers1-3. It has been established that whole-genome duplications fuel chromosome instability through abnormal mitosis4-8; however, the immediate consequences of tetraploidy in the first interphase are not known. This is a key question because single whole-genome duplication events such as cytokinesis failure can promote tumorigenesis9. Here we find that human cells undergo high rates of DNA damage during DNA replication in the first S phase following induction of tetraploidy. Using DNA combing and single-cell sequencing, we show that DNA replication dynamics is perturbed, generating under- and over-replicated regions. Mechanistically, we find that these defects result from a shortage of proteins during the G1/S transition, which impairs the fidelity of DNA replication. This work shows that within a single interphase, unscheduled tetraploid cells can acquire highly abnormal karyotypes. These findings provide an explanation for the genetic instability landscape that favours tumorigenesis after tetraploidization.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










Comment in
-
Genome doubling causes double trouble.Nature. 2022 Apr;604(7904):44-45. doi: 10.1038/d41586-022-00849-2. Nature. 2022. PMID: 35354975 No abstract available.
-
Catastrophic DNA replication in unscheduled tetraploid cells.Trends Genet. 2022 Aug;38(8):787-788. doi: 10.1016/j.tig.2022.04.005. Epub 2022 Apr 27. Trends Genet. 2022. PMID: 35490031
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

