Human distal lung maps and lineage hierarchies reveal a bipotent progenitor
- PMID: 35355018
- PMCID: PMC9169066
- DOI: 10.1038/s41586-022-04541-3
Human distal lung maps and lineage hierarchies reveal a bipotent progenitor
Abstract
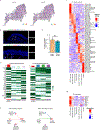
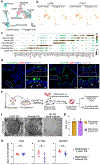
Mapping the spatial distribution and molecular identity of constituent cells is essential for understanding tissue dynamics in health and disease. We lack a comprehensive map of human distal airways, including the terminal and respiratory bronchioles (TRBs), which are implicated in respiratory diseases1-4. Here, using spatial transcriptomics and single-cell profiling of microdissected distal airways, we identify molecularly distinct TRB cell types that have not-to our knowledge-been previously characterized. These include airway-associated LGR5+ fibroblasts and TRB-specific alveolar type-0 (AT0) cells and TRB secretory cells (TRB-SCs). Connectome maps and organoid-based co-cultures reveal that LGR5+ fibroblasts form a signalling hub in the airway niche. AT0 cells and TRB-SCs are conserved in primates and emerge dynamically during human lung development. Using a non-human primate model of lung injury, together with human organoids and tissue specimens, we show that alveolar type-2 cells in regenerating lungs transiently acquire an AT0 state from which they can differentiate into either alveolar type-1 cells or TRB-SCs. This differentiation programme is distinct from that identified in the mouse lung5-7. Our study also reveals mechanisms that drive the differentiation of the bipotent AT0 cell state into normal or pathological states. In sum, our findings revise human lung cell maps and lineage trajectories, and implicate an epithelial transitional state in primate lung regeneration and disease.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interest declaration
P.R.T. serves as a consultant for Surrozen Inc., Cellarity Inc., and Celldom Inc., on work not related to the contents of this manuscript. JRR is an employee of Genentech Inc. The other authors declare no competing interests.
Figures















References
-
- Hogg JC, Macklem PT & Thurlbeck WM The resistance of small airways in normal and diseased human lungs. Aspen Emphysema Conf 10, 433–441 (1967). - PubMed
-
- Weibel ER & Gomez DM Architecture of the human lung. Use of quantitative methods establishes fundamental relations between size and number of lung structures. Science 137, 577–585 (1962). - PubMed
MeSH terms
Grants and funding
- R01 HL119215/HL/NHLBI NIH HHS/United States
- R01 HL153375/HL/NHLBI NIH HHS/United States
- R00 HL127181/HL/NHLBI NIH HHS/United States
- R24 HD000836/HD/NICHD NIH HHS/United States
- R01 HL165404/HL/NHLBI NIH HHS/United States
- P51 OD011107/OD/NIH HHS/United States
- R01 HL127002/HL/NHLBI NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
- U01 HL134766/HL/NHLBI NIH HHS/United States
- R01 HL166139/HL/NHLBI NIH HHS/United States
- R01 DK047967/DK/NIDDK NIH HHS/United States
- R01 HL146557/HL/NHLBI NIH HHS/United States
- P30 DK065988/DK/NIDDK NIH HHS/United States
- T32 GM007315/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

