Tap water as a natural vehicle for microorganisms shaping the human gut microbiome
- PMID: 35355372
- PMCID: PMC9790288
- DOI: 10.1111/1462-2920.15988
Tap water as a natural vehicle for microorganisms shaping the human gut microbiome
Abstract
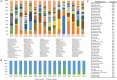
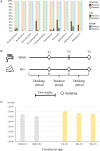
Fresh potable water is an indispensable drink which humans consume daily in substantial amounts. Nonetheless, very little is known about the composition of the microbial community inhabiting drinking water or its impact on our gut microbiota. In the current study, an exhaustive shotgun metagenomics analysis of the tap water microbiome highlighted the occurrence of a highly genetic biodiversity of the microbial communities residing in fresh water and the existence of a conserved core tap water microbiota largely represented by novel microbial species, representing microbial dark matter. Furthermore, genome reconstruction of this microbial dark matter from water samples unveiled homologous sequences present in the faecal microbiome of humans from various geographical locations. Accordingly, investigation of the faecal microbiota content of a subject that daily consumed tap water for 3 years provides proof for horizontal transmission and colonization of water bacteria in the human gut.
© 2022 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures

References
-
- Bowyer, R.C.E. , Schillereff, D.N. , Jackson, M.A. , Le Roy, C. , Wells, p.M. , Spector, T.D. , and Steves, C.J. (2020) Associations between UKtap water and gut microbiota composition suggest the gut microbiome as a potential mediator of health differences linked to water quality. Sci Total Environ 739: 139697. - PubMed
-
- Busse, H.J. , Denner, E.B.M. , Buczolits, S. , Salkinoja‐Salonen, M. , Bennasar, A. , and Kämpfer, P. (2003) Sphingomonas aurantiaca sp. nov., Sphingomonas aerolata sp. nov. and Sphingomonas faeni sp. nov., air‐ and dustborne and Antarctic, orange‐pigmented, psychrotolerant bacteria, and emended description of the genus Sphingomonas. Int J Syst Evol Microbiol 53: 1253–1260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

