Molecular insights into the biochemical functions and signalling mechanisms of plant NLRs
- PMID: 35355394
- PMCID: PMC9104254
- DOI: 10.1111/mpp.13195
Molecular insights into the biochemical functions and signalling mechanisms of plant NLRs
Abstract
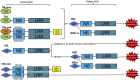
Plant intracellular immune receptors known as NLR (nucleotide-binding leucine-rich repeat) proteins confer immunity and cause cell death. Plant NLR proteins that directly or indirectly recognize pathogen effector proteins to initiate immune signalling are regarded as sensor NLRs. Some NLR protein families function downstream of sensor NLRs to transduce immune signalling and are known as helper NLRs. Recent breakthrough studies on plant NLR protein structures and biochemical functions greatly advanced our understanding of NLR biology. Comprehensive and detailed knowledge on NLR biology requires future efforts to solve more NLR protein structures and investigate the signalling events between sensor and helper NLRs, and downstream of helper NLRs.
Keywords: cell death; immune receptors; pathogen effector; plant immunity; signal transduction.
© 2022 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures



Similar articles
-
Resistosomes at the interface of pathogens and plants.Curr Opin Plant Biol. 2022 Jun;67:102212. doi: 10.1016/j.pbi.2022.102212. Epub 2022 Apr 21. Curr Opin Plant Biol. 2022. PMID: 35462196 Review.
-
Direct recognition of pathogen effectors by plant NLR immune receptors and downstream signalling.Essays Biochem. 2022 Sep 30;66(5):471-483. doi: 10.1042/EBC20210072. Essays Biochem. 2022. PMID: 35731245 Free PMC article. Review.
-
Effector-dependent activation and oligomerization of plant NRC class helper NLRs by sensor NLR immune receptors Rpi-amr3 and Rpi-amr1.EMBO J. 2023 Mar 1;42(5):e111484. doi: 10.15252/embj.2022111484. Epub 2023 Jan 2. EMBO J. 2023. PMID: 36592032 Free PMC article.
-
Interfamily co-transfer of sensor and helper NLRs extends immune receptor functionality between angiosperms.Cell. 2025 Aug 21;188(17):4505-4516.e14. doi: 10.1016/j.cell.2025.05.028. Epub 2025 Jun 17. Cell. 2025. PMID: 40532698
-
Animal NLRs provide structural insights into plant NLR function.Ann Bot. 2017 Mar 1;119(5):827-702. doi: 10.1093/aob/mcw171. Ann Bot. 2017. PMID: 27562749 Free PMC article. Review.
Cited by
-
Activation and Autoinhibition Mechanisms of NLR Immune Receptor Pi36 in Rice.Int J Mol Sci. 2024 Jul 2;25(13):7301. doi: 10.3390/ijms25137301. Int J Mol Sci. 2024. PMID: 39000408 Free PMC article.
-
A catalogue of virulence strategies mediated by phytopathogenic effectors.Fundam Res. 2024 Feb 21;5(2):663-673. doi: 10.1016/j.fmre.2023.10.026. eCollection 2025 Mar. Fundam Res. 2024. PMID: 40242527 Free PMC article. Review.
-
Activation and inhibition mechanisms of a plant helper NLR.Nature. 2025 Mar;639(8054):438-446. doi: 10.1038/s41586-024-08517-3. Epub 2025 Feb 12. Nature. 2025. PMID: 39939758
-
Plasma membrane association and resistosome formation of plant helper immune receptors.Proc Natl Acad Sci U S A. 2023 Aug 8;120(32):e2222036120. doi: 10.1073/pnas.2222036120. Epub 2023 Jul 31. Proc Natl Acad Sci U S A. 2023. PMID: 37523563 Free PMC article.
-
Ferroptosis, necroptosis, and pyroptosis in the occurrence and development of ovarian cancer.Front Immunol. 2022 Jul 25;13:920059. doi: 10.3389/fimmu.2022.920059. eCollection 2022. Front Immunol. 2022. PMID: 35958626 Free PMC article. Review.
References
-
- Adachi, H. , Derevnina, L. & Kamoun, S. (2019) NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Current Opinion in Plant Biology, 50, 121–131. - PubMed
-
- Bi, G. , Su, M. , Li, N. , Liang, Y.U. , Dang, S. , Xu, J. et al. (2021) The ZAR1 resistosome is a calcium‐permeable channel triggering plant immune signaling. Cell, 184, 3528–3541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

