Gut microbiome in liver pathophysiology and cholestatic liver disease
- PMID: 35355516
- PMCID: PMC8963136
- DOI: 10.1016/j.livres.2021.08.001
Gut microbiome in liver pathophysiology and cholestatic liver disease
Abstract
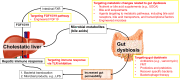
An increasing amount of evidence has shown critical roles of gut microbiome in host pathophysiology. The gut and the liver are anatomically and physiologically connected. Given the critical role of gut-liver axis in the homeostasis of the liver, gut microbiome interplays with a diverse spectrum of hepatic changes, including steatosis, inflammation, fibrosis, cholestasis, and tumorigenesis. In clinic, cholestasis manifests with fatigue, pruritus, and jaundice, caused by the impairment in bile formation or flow. Studies have shown that the gut microbiome is altered in cholestatic liver disease. In this review, we will explore the interaction between the gut microbiome and the liver with a focus on the alteration and the role of gut microbiome in cholestatic liver disease. We will also discuss the prospect of exploiting the gut microbiome in the development of novel therapies for cholestatic liver disease.
Keywords: Cholestatic liver disease; Gut microbiome; Liver pathophysiology.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no conflict of interest.
Figures
Similar articles
-
The gut microbial influence on cholestatic liver disease.Liver Int. 2019 Jul;39(7):1186-1196. doi: 10.1111/liv.14153. Epub 2019 Jun 17. Liver Int. 2019. PMID: 31125502 Review.
-
The Gut-Liver Axis in Cholestatic Liver Diseases.Nutrients. 2021 Mar 21;13(3):1018. doi: 10.3390/nu13031018. Nutrients. 2021. PMID: 33801133 Free PMC article. Review.
-
Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases.Autoimmun Rev. 2017 Sep;16(9):885-896. doi: 10.1016/j.autrev.2017.07.002. Epub 2017 Jul 8. Autoimmun Rev. 2017. PMID: 28698093 Review.
-
Gut-Liver Axis and Inflammasome Activation in Cholangiocyte Pathophysiology.Cells. 2020 Mar 17;9(3):736. doi: 10.3390/cells9030736. Cells. 2020. PMID: 32192118 Free PMC article. Review.
-
Intestinal microbiota drives cholestasis-induced specific hepatic gene expression patterns.Gut Microbes. 2021 Jan-Dec;13(1):1-20. doi: 10.1080/19490976.2021.1911534. Gut Microbes. 2021. PMID: 33847205 Free PMC article.
Cited by
-
Bile acid metabolism and bile acid receptor signaling in metabolic diseases and therapy.Liver Res. 2021 Aug 28;5(3):103-104. doi: 10.1016/j.livres.2021.08.002. eCollection 2021 Sep. Liver Res. 2021. PMID: 39957844 Free PMC article. No abstract available.
-
An untargeted comparative metabolomics analysis of infants with and without late-onset breast milk jaundice.PLoS One. 2024 Aug 12;19(8):e0308710. doi: 10.1371/journal.pone.0308710. eCollection 2024. PLoS One. 2024. PMID: 39133689 Free PMC article.
-
Antibiotics in Chronic Liver Disease and Their Effects on Gut Microbiota.Antibiotics (Basel). 2023 Sep 22;12(10):1475. doi: 10.3390/antibiotics12101475. Antibiotics (Basel). 2023. PMID: 37887176 Free PMC article. Review.
-
Hepatoprotective Properties of Water Kefir: A Traditional Fermented Drink and Its Potential Role.Int J Prev Med. 2023 Jul 15;14:93. doi: 10.4103/ijpvm.ijpvm_29_22. eCollection 2023. Int J Prev Med. 2023. PMID: 37855014 Free PMC article. Review.
-
Alleviation of cholestatic liver injury and intestinal permeability by lubiprostone treatment in bile duct ligated rats: role of intestinal FXR and tight junction proteins claudin-1, claudin-2, and occludin.Naunyn Schmiedebergs Arch Pharmacol. 2023 Sep;396(9):2009-2022. doi: 10.1007/s00210-023-02455-z. Epub 2023 Mar 10. Naunyn Schmiedebergs Arch Pharmacol. 2023. PMID: 36897372
References
Grants and funding
LinkOut - more resources
Full Text Sources
