A Shaving Proteomic Approach to Unveil Surface Proteins Modulation of Multi-Drug Resistant Pseudomonas aeruginosa Strains Isolated From Cystic Fibrosis Patients
- PMID: 35355602
- PMCID: PMC8959810
- DOI: 10.3389/fmed.2022.818669
A Shaving Proteomic Approach to Unveil Surface Proteins Modulation of Multi-Drug Resistant Pseudomonas aeruginosa Strains Isolated From Cystic Fibrosis Patients
Abstract
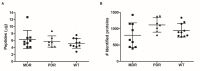
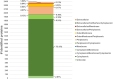
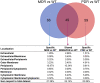
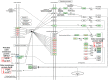
Cystic fibrosis (CF) is the most common rare disease caused by a mutation of the CF transmembrane conductance regulator gene encoding a channel protein of the apical membrane of epithelial cells leading to alteration of Na+ and K+ transport, hence inducing accumulation of dense and sticky mucus and promoting recurrent airway infections. The most detected bacterium in CF patients is Pseudomonas aeruginosa (PA) which causes chronic colonization, requiring stringent antibiotic therapies that, in turn induces multi-drug resistance. Despite eradication attempts at the first infection, the bacterium is able to utilize several adaptation mechanisms to survive in hostile environments such as the CF lung. Its adaptive machinery includes modulation of surface molecules such as efflux pumps, flagellum, pili and other virulence factors. In the present study we compared surface protein expression of PA multi- and pan-drug resistant strains to wild-type antibiotic-sensitive strains, isolated from the airways of CF patients with chronic colonization and recent infection, respectively. After shaving with trypsin, microbial peptides were analyzed by tandem-mass spectrometry on a high-resolution platform that allowed the identification of 174 differentially modulated proteins localized in the region from extracellular space to cytoplasmic membrane. Biofilm assay was performed to characterize all 26 PA strains in term of biofilm production. Among the differentially expressed proteins, 17 were associated to the virulome (e.g., Tse2, Tse5, Tsi1, PilF, FliY, B-type flagellin, FliM, PyoS5), six to the resistome (e.g., OprJ, LptD) and five to the biofilm reservoir (e.g., AlgF, PlsD). The biofilm assay characterized chronic antibiotic-resistant isolates as weaker biofilm producers than wild-type strains. Our results suggest the loss of PA early virulence factors (e.g., pili and flagella) and later expression of virulence traits (e.g., secretion systems proteins) as an indicator of PA adaptation and persistence in the CF lung environment. To our knowledge, this is the first study that, applying a shaving proteomic approach, describes adaptation processes of a large collection of PA clinical strains isolated from CF patients in early and chronic infection phases.
Keywords: Pseudomonas aeruginosa; antibiotic resistance; cystic fibrosis; long-term colonization; shaving proteomics.
Copyright © 2022 Montemari, Marzano, Essa, Levi Mortera, Rossitto, Gardini, Selan, Vrenna, Onetti Muda, Putignani and Fiscarelli.
Conflict of interest statement
SG was employed by GenomeUp. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Mortensen KL, Jensen RH, Johansen HK, Skov M, Pressler T, Howard SJ, et al. Aspergillus species and other molds in respiratory samples from patients with cystic fibrosis: a laboratory-based study with focus on Aspergillus fumigatus azole resistance. J Clin Microbiol. (2011) 49:2243–51. 10.1128/JCM.00213-11 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

