The Heterogeneity of Immune Cell Infiltration Landscape and Its Immunotherapeutic Implications in Hepatocellular Carcinoma
- PMID: 35355983
- PMCID: PMC8959995
- DOI: 10.3389/fimmu.2022.861525
The Heterogeneity of Immune Cell Infiltration Landscape and Its Immunotherapeutic Implications in Hepatocellular Carcinoma
Abstract
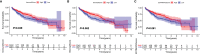
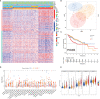
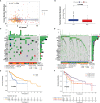
Immunotherapy, closely associated with immune infiltration and tumor mutation burden (TMB), is emerging as a promising strategy for treating tumors, but its low response rate in hepatocellular carcinoma (HCC) remains a major challenge. Herein, we applied two algorithms to uncover the immune infiltration landscape of the immune microenvironment in 491 HCC patients. Three immune infiltration patterns were defined using the CIBERSORT method, and the immune cell infiltration (ICI) scores were established using principal component analysis. In the high ICI score group, the activation of the Wnt/β-catenin pathway was significantly enriched and expressions of immune checkpoint genes increased, which showed a pessimistic outcome. The low ICI score group was characterized by increased TMB and enrichment of metabolism-related pathways. Further analysis found that the ICI score exhibited a significant difference in age ≥65/age <65, grade I/grade II-IV, and response to immunotherapy. Moreover, the CTNNB1 mutation status was found to be closely associated with prognosis and immunotherapeutic efficiency, significantly affecting the ICI score and TMB, which might be regarded as a potential marker for the treatment of HCC. The evaluation of immune infiltration patterns can improve the understanding of the tumor immune microenvironment and provide new directions for the study of individualized immunotherapy strategies for HCC.
Keywords: hepatocellular carcinoma; immune cell infiltration; immune microenvironment; tumor heterogeneity; tumor mutational burden.
Copyright © 2022 Guo, Yang, Ren, Tian, Gao, Tian, Zhang and Kan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Osipov A, Lim SJ, Popovic A, Azad NS, Laheru DA, Zheng L, et al. Tumor Mutational Burden, Toxicity, and Response of Immune Checkpoint Inhibitors Targeting PD(L)1, CTLA-4, and Combination: A Meta-Regression Analysis. Clin Cancer Res (2020) 26:4842–51. doi: 10.1158/1078-0432.CCR-20-0458 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

