Response of Human Liver Tissue to Innate Immune Stimuli
- PMID: 35355993
- PMCID: PMC8959492
- DOI: 10.3389/fimmu.2022.811551
Response of Human Liver Tissue to Innate Immune Stimuli
Abstract
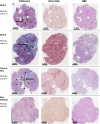
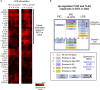
Precision-cut human liver slice cultures (PCLS) have become an important alternative immunological platform in preclinical testing. To further evaluate the capacity of PCLS, we investigated the innate immune response to TLR3 agonist (poly-I:C) and TLR4 agonist (LPS) using normal and diseased liver tissue. Pathological liver tissue was obtained from patients with active chronic HCV infection, and patients with former chronic HCV infection cured by recent Direct-Acting Antiviral (DAA) drug therapy. We found that hepatic innate immunity in response to TLR3 and TLR4 agonists was not suppressed but enhanced in the HCV-infected tissue, compared with the healthy controls. Furthermore, despite recent HCV elimination, DAA-cured liver tissue manifested ongoing abnormalities in liver immunity: sustained abnormal immune gene expression in DAA-cured samples was identified in direct ex vivo measurements and in TLR3 and TLR4 stimulation assays. Genes that were up-regulated in chronic HCV-infected liver tissue were mostly characteristic of the non-parenchymal cell compartment. These results demonstrated the utility of PCLS in studying both liver pathology and innate immunity.
Keywords: direct-acting antiviral treatment; fibrosis; hepatitis C virus; inflammation; innate immunity; liver slice culture.
Copyright © 2022 Wu, Roberto, Knupp, Greninger, Truong, Hollingshead, Kenerson, Tuefferd, Chen, Koelle, Horton, Jerome, Polyak, Yeung and Crispe.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

