Staphylococcus aureus Multiplexes Death-Effector Deoxyribonucleosides to Neutralize Phagocytes
- PMID: 35355997
- PMCID: PMC8960049
- DOI: 10.3389/fimmu.2022.847171
Staphylococcus aureus Multiplexes Death-Effector Deoxyribonucleosides to Neutralize Phagocytes
Abstract
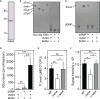
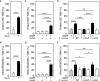
Adenosine synthase A (AdsA) is a key virulence factor of Staphylococcus aureus, a dangerous microbe that causes fatal diseases in humans. Together with staphylococcal nuclease, AdsA generates deoxyadenosine (dAdo) from neutrophil extracellular DNA traps thereby igniting caspase-3-dependent cell death in host immune cells that aim at penetrating infectious foci. Powered by a multi-technological approach, we here illustrate that the enzymatic activity of AdsA in abscess-mimicking microenvironments is not restricted to the biogenesis of dAdo but rather comprises excessive biosynthesis of deoxyguanosine (dGuo), a cytotoxic deoxyribonucleoside generated by S. aureus to eradicate macrophages of human and animal origin. Based on a genome-wide CRISPR-Cas9 knock-out screen, we further demonstrate that dGuo-induced cytotoxicity in phagocytes involves targeting of the mammalian purine salvage pathway-apoptosis axis, a signaling cascade that is concomitantly stimulated by staphylococcal dAdo. Strikingly, synchronous targeting of this route by AdsA-derived dGuo and dAdo boosts macrophage cell death, indicating that S. aureus multiplexes death-effector deoxyribonucleosides to maximize intra-host survival. Overall, these data provide unique insights into the cunning lifestyle of a deadly pathogen and may help to design therapeutic intervention strategies to combat multidrug-resistant staphylococci.
Keywords: Staphylococcus aureus; apoptosis; deoxyguanosine; deoxyribonucleosides; immune evasion; macrophage; phagocyte.
Copyright © 2022 Tantawy, Schwermann, Ostermeier, Garbe, Bähre, Vital and Winstel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

