A scalable serology solution for profiling humoral immune responses to SARS-CoV-2 infection and vaccination
- PMID: 35356067
- PMCID: PMC8942165
- DOI: 10.1002/cti2.1380
A scalable serology solution for profiling humoral immune responses to SARS-CoV-2 infection and vaccination
Abstract
Objectives: Antibody testing against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been instrumental in detecting previous exposures and analyzing vaccine-elicited immune responses. Here, we describe a scalable solution to detect and quantify SARS-CoV-2 antibodies, discriminate between natural infection- and vaccination-induced responses, and assess antibody-mediated inhibition of the spike-angiotensin converting enzyme 2 (ACE2) interaction.
Methods: We developed methods and reagents to detect SARS-CoV-2 antibodies by enzyme-linked immunosorbent assay (ELISA). The main assays focus on the parallel detection of immunoglobulin (Ig)Gs against the spike trimer, its receptor binding domain (RBD) and nucleocapsid (N). We automated a surrogate neutralisation (sn)ELISA that measures inhibition of ACE2-spike or -RBD interactions by antibodies. The assays were calibrated to a World Health Organization reference standard.
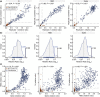
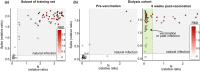
Results: Our single-point IgG-based ELISAs accurately distinguished non-infected and infected individuals. For seroprevalence assessment (in a non-vaccinated cohort), classifying a sample as positive if antibodies were detected for ≥ 2 of the 3 antigens provided the highest specificity. In vaccinated cohorts, increases in anti-spike and -RBD (but not -N) antibodies are observed. We present detailed protocols for serum/plasma or dried blood spots analysis performed manually and on automated platforms. The snELISA can be performed automatically at single points, increasing its scalability.
Conclusions: Measuring antibodies to three viral antigens and identify neutralising antibodies capable of disrupting spike-ACE2 interactions in high-throughput enables large-scale analyses of humoral immune responses to SARS-CoV-2 infection and vaccination. The reagents are available to enable scaling up of standardised serological assays, permitting inter-laboratory data comparison and aggregation.
Keywords: SARS‐CoV‐2; antibody detection; antibody neutralisation; assay development and standardisation; high‐throughput screening.
© 2022 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Johns Hopkins University COVID‐19 dashboard. [cited February 25 2021]. Available from: https://coronavirus.jhu.edu/map.html
-
- Few Canadians had antibodies against SARS‐CoV‐2 in early 2021 – Statistics Canada. [updated July 6 2021; cited February 25 2022]. Available from: https://www150.statcan.gc.ca/n1/daily‐quotidien/210706/dq210706a‐eng.htm
-
- Serotracker. [updated February 22 2022; cited February 25 2022]. Available from: https://serotracker.com/en/Explore
-
- Health Infobase Canada – COVID‐19 vaccination coverage. [updated February 18 2022; cited February 25 2022]. Available from: https://health‐infobase.canada.ca/covid‐19/vaccination‐coverage/
LinkOut - more resources
Full Text Sources
Miscellaneous
