Prolonged Outbreak of Carbapenem and Colistin-Resistant Klebsiella pneumoniae at a Large Tertiary Hospital in Brazil
- PMID: 35356529
- PMCID: PMC8959819
- DOI: 10.3389/fmicb.2022.831770
Prolonged Outbreak of Carbapenem and Colistin-Resistant Klebsiella pneumoniae at a Large Tertiary Hospital in Brazil
Abstract
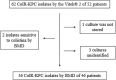
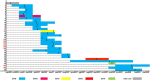
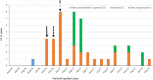
Multidrug-resistant gram-negative bacteria, such as carbapenem and colistin-resistant Klebsiella pneumoniae (ColR-CRKP), represent a major problem for health systems worldwide and have high lethality. This study investigated the genetic relationship, antimicrobial susceptibility profile, and resistance mechanisms to ColR-CRKP isolates from patients infected/colonized in a tertiary hospital in Salvador, Bahia/Brazil. From September 2016 to January 2018, 46 patients (56 ColR-CRKP positive cultures) were enrolled in the investigation but clinical and demographic data were obtained from 31 patients. Most of them were men (67.7%) and elderly (median age of 62 years old), and the median Charlson score was 3. The main comorbidities were systemic arterial hypertension (38.7%), diabetes (32.2%), and cerebrovascular disease (25.8%). The average hospitalization stay until ColR-CRKP identification in days were 35.12. A total of 90.6% used mechanical ventilation and 93.7% used a central venous catheter. Of the 31 patients who had the data evaluated, 12 had ColR-CRKP infection, and seven died (58.4%). Previous use of polymyxins was identified in 32.2% of the cases, and carbapenems were identified in 70.9%. The minimum inhibitory concentration (MIC) for colistin was > 16 μg/mL, with more than half of the isolates (55%) having a MIC of 256 μg/mL. The bla KPC gene was detected in 94.7% of the isolates, bla NDM in 16.0%, and bla GES in 1.7%. The bla OXA-48, bla VIM, and bla IMP genes were not detected. The mcr-1 test was negative in all 56 isolates. Alteration of the mgrB gene was detected in 87.5% (n = 49/56) of the isolates, and of these, 49.0% (24/49) had alteration in size probably due to IS903B, 22.4% (11/49) did not have the mgrB gene detected, 20.4% (10/49) presented the IS903B, 6.1% (3/49) had a premature stop codon (Q30*), and 2.1% (1/49) presented a thymine deletion at position 104 - 104delT (F35fs). The PFGE profile showed a monoclonal profile in 84.7% of the isolates in different hospital sectors, with ST11 (CC-258) being the most frequent sequence type. This study presents a prolonged outbreak of ColR-CRKP in which 83.9% of the isolates belonged to the same cluster, and 67.6% of the patients evaluated had not used polymyxin, suggesting the possibility of cross-transmission of ColR-CRKP isolates.
Keywords: Enterobacterales infections; carbapenem-resistant Klebsiella pneumoniae; colistin-resistant Klebsiella pneumoniae; multidrug-resistant gram-negative bacteria; outbreak.
Copyright © 2022 Rocha, Barbosa, Leal, Silva, Sales, Monteiro, Azevedo, Malheiros, Ataide, Moreira, Reis, Bahia and Reis.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Aires C. A. M., Conceição-Neto O. C., Oliveira T. R. T., Dias C. F., Montezzi L. F., Picão R. C., et al. (2017). Emergence of the plasmid-mediated mcr-1 gene in clinical KPC-2-producing Klebsiella pneumoniae sequence type 392 in Brazil. Antimicrob. Agents Chemother. 61 e00317–17. 10.1128/AAC.00317-17 - DOI - PMC - PubMed
-
- Aulin L. B. S., Koumans C. I. M., Haakman Y., van Os W., Kraakman M. E. M., Gooskens J., et al. (2021). Distinct evolution of colistin resistance associated with experimental resistance evolution models in Klebsiella pneumoniae. J. Antimicrob. Chemother. 76 533–535. 10.1093/jac/dkaa450 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

