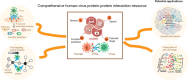
Comprehensive characterization of human-virus protein-protein interactions reveals disease comorbidities and potential antiviral drugs
- PMID: 35356543
- PMCID: PMC8924640
- DOI: 10.1016/j.csbj.2022.03.002
Comprehensive characterization of human-virus protein-protein interactions reveals disease comorbidities and potential antiviral drugs
Abstract
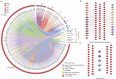
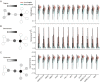
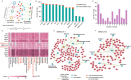
The protein-protein interactions (PPIs) between human and viruses play important roles in viral infection and host immune responses. Rapid accumulation of experimentally validated human-virus PPIs provides an unprecedented opportunity to investigate the regulatory pattern of viral infection. However, we are still lack of knowledge about the regulatory patterns of human-virus interactions. We collected 27,293 experimentally validated human-virus PPIs, covering 8 virus families, 140 viral proteins and 6059 human proteins. Functional enrichment analysis revealed that the viral interacting proteins were likely to be enriched in cell cycle and immune-related pathways. Moreover, we analysed the topological features of the viral interacting proteins and found that they were likely to locate in central regions of human PPI network. Based on network proximity analyses of diseases genes and human-virus interactions in the human interactome, we revealed the associations between complex diseases and viral infections. Network analysis also implicated potential antiviral drugs that were further validated by text mining. Finally, we presented the Human-Virus Protein-Protein Interaction database (HVPPI, http://bio-bigdata.hrbmu.edu.cn/HVPPI), that provides experimentally validated human-virus PPIs as well as seamlessly integrates online functional analysis tools. In summary, comprehensive understanding the regulatory pattern of human-virus interactome will provide novel insights into fundamental infectious mechanism discovery and new antiviral therapy development.
Keywords: Antiviral therapy; Disease comorbidities; HVPPI; Network analysis; Protein-protein interactions.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
HVIDB: a comprehensive database for human-virus protein-protein interactions.Brief Bioinform. 2021 Mar 22;22(2):832-844. doi: 10.1093/bib/bbaa425. Brief Bioinform. 2021. PMID: 33515030
-
Understanding Human-Virus Protein-Protein Interactions Using a Human Protein Complex-Based Analysis Framework.mSystems. 2019 Apr 9;4(2):e00303-18. doi: 10.1128/mSystems.00303-18. eCollection 2019 Mar-Apr. mSystems. 2019. PMID: 30984872 Free PMC article.
-
Current status and future perspectives of computational studies on human-virus protein-protein interactions.Brief Bioinform. 2021 Sep 2;22(5):bbab029. doi: 10.1093/bib/bbab029. Brief Bioinform. 2021. PMID: 33693490 Review.
-
Multi-omics characterization of RNA binding proteins reveals disease comorbidities and potential drugs in COVID-19.Comput Biol Med. 2023 Mar;155:106651. doi: 10.1016/j.compbiomed.2023.106651. Epub 2023 Feb 10. Comput Biol Med. 2023. PMID: 36805221 Free PMC article.
-
Mapping the SARS-CoV-2-Host Protein-Protein Interactome by Affinity Purification Mass Spectrometry and Proximity-Dependent Biotin Labeling: A Rational and Straightforward Route to Discover Host-Directed Anti-SARS-CoV-2 Therapeutics.Int J Mol Sci. 2021 Jan 7;22(2):532. doi: 10.3390/ijms22020532. Int J Mol Sci. 2021. PMID: 33430309 Free PMC article. Review.
Cited by
-
Prediction of human protein interactome of dengue virus non-structural protein 5 (NS5) and its downstream immunological implications.3 Biotech. 2023 Jun;13(6):180. doi: 10.1007/s13205-023-03569-0. Epub 2023 May 12. 3 Biotech. 2023. PMID: 37193327 Free PMC article.
-
Malivhu: A Comprehensive Bioinformatics Resource for Filtering SARS and MERS Virus Proteins by Their Classification, Family and Species, and Prediction of Their Interactions Against Human Proteins.Bioinform Biol Insights. 2024 Aug 14;18:11779322241263671. doi: 10.1177/11779322241263671. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39148721 Free PMC article.
-
Exploring Viral-Host Protein Interactions as Antiviral Therapies: A Computational Perspective.Microorganisms. 2024 Mar 21;12(3):630. doi: 10.3390/microorganisms12030630. Microorganisms. 2024. PMID: 38543681 Free PMC article. Review.
-
Integrating multi-omics to unravel host-microbiome interactions in inflammatory bowel disease.Cell Rep Med. 2024 Sep 17;5(9):101738. doi: 10.1016/j.xcrm.2024.101738. Cell Rep Med. 2024. PMID: 39293401 Free PMC article. Review.
-
Proteome-wide characterization of PTMs reveals host cell responses to viral infection and identifies putative antiviral drug targets.Front Immunol. 2025 May 30;16:1587106. doi: 10.3389/fimmu.2025.1587106. eCollection 2025. Front Immunol. 2025. PMID: 40519923 Free PMC article. Review.
References
-
- Lian X., Yang X., Yang S., Zhang Z. Current status and future perspectives of computational studies on human-virus protein-protein interactions. Brief Bioinform. 2021 - PubMed
LinkOut - more resources
Full Text Sources
